Single-cell RNA seq method developed to accurately quantify cell-specific drug effects in pancreatic islets
Findings could open up potential therapeutic avenues to treat Type 1 diabetes
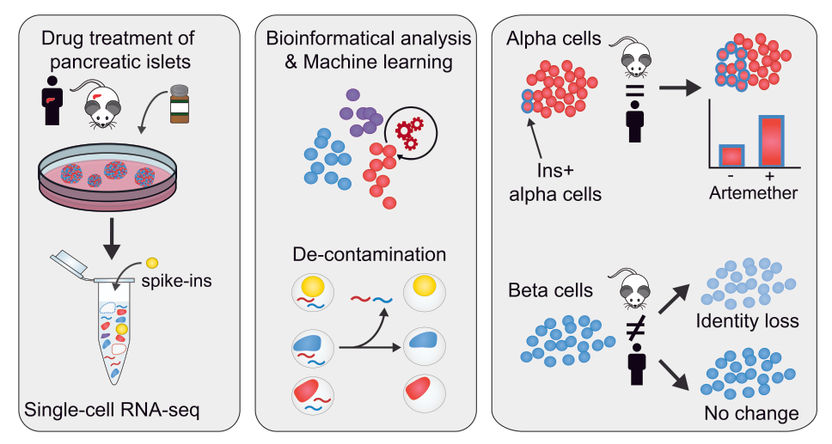
Researchers from Stefan Kubicek’s and Christoph Bock’s groups, at the CeMM Research Center for Molecular Medicine of the Austrian Academy of Sciences in Vienna, have developed a method to accurately assess the effect of specific drugs in isolated pancreatic tissue by using a refined single-cell RNA sequencing method. Their study published in Genome Biology describes the technique that they have developed to overcome the problem of contaminating RNA molecules in single-cell transcriptomics, which allowed for accurate results of dynamic drug responses in pancreatic cells. These findings will support the development of targeted drug therapies for the treatment of Type 1 diabetes in the future.
Graphical abstract
B. Marquina-Sánchez N. Fortelny
The pancreas is an abdominal organ that produces digestive enzymes as well as hormones that regulate blood sugar levels. This hormone-producing function is localized to the islets of Langerhans, which constitute clusters of different endocrine cell types. Among those are beta cells, which produce the hormone insulin needed to lower glucose (a type of sugar) levels in our blood, as well as alpha cells, which generate the hormone glucagon in charge of raising glucose levels in the blood.
Type 1 diabetes is a chronic disease in which the body’s immune system mistakenly attacks and destroys the pancreas’ insulin-producing beta cells. Regenerative medicine aims to replenish beta cell mass, and thus support and ultimately substitute the current insulin replacement therapies. Alterations to islet composition, including insufficient beta cell function and beta cell dedifferention, also contribute to type II diabetes. Therefore, a deeper understanding of the identity and crosstalk of the different islet cell types leads to a better characterization of both forms of diabetes and may contribute to the development of novel therapeutic concepts.
Single-cell transcriptomics is a powerful technique to characterize cellular identity. Previously, CeMM researchers from Christoph Bock’s and Stefan Kubicek’s groups at CeMM published the first single cell transcriptomes from primary human pancreatic islet cells (EMBO Rep. 2016 Feb 17;(2):178-87). Advances in the technology have since enabled its application to the generation of global human and mouse single cell transcriptome atlases. Despite these advances, single cell approaches remain technologically challenging given that the miniscule RNA amount present is entirely used up in the experiment. Therefore, it is essential to ensure the quality and purity of the resulting single cell transcriptomes.
CeMM researchers in the two contributing laboratories identified unexpectedly high hormone expression in non-endocrine cell types, both in their own dataset as well as other published single cell studies. They set out to elucidate whether this would be the result of contamination by RNA molecules, for example from dying cells, and how it could be removed to obtain a more reliable dataset. Such contamination seems present in single cell RNA-seq data from most tissues but was most visible in pancreatic islets. Islet endocrine cells are exclusively devoted to the production of single hormones, and insulin in beta cells and glucagon in alpha cells are expressed to higher levels than typical “housekeeping” genes. Thus, redistribution of these transcripts to other cell types was highly pronounced. Based on this observation, their goal was to develop, validate and apply a method to experimentally determine and computationally remove such contamination.
In their investigation, CeMM researchers used spiked-in cells from different cell types, both mouse and human samples, that they added to their pancreatic islet samples. Importantly, the transcriptomes of these spike-in cell were fully characterized. This allowed them to control internally and accurately the level of RNA contamination in single cell RNA-seq, giving that the human transcripts detected in the mouse spike-in cells constitute contaminating RNA. In this way, they found that the samples had a contamination level of up to 20%, and were able to define the contamination in each samples. They then developed a novel bioinformatics approach to computationally remove contaminating reads from single cell transcriptomes.
Having now obtained a “decontaminated” transcriptome, from which the spurious signal has been removed, they proceeded to characterize how the cellular identity in the different cell types responded to the treatment with three different drugs. They found that a small molecule inhibitor of the transcription factor FOXO1 induces dedifferentiation of both alpha and beta cells. Furthermore, they studied artemether, which had been found to diminish the function of alpha cells and could induce insulin production in both in vivo and in vitro studies (Cell. 2017 Jan 12;168(1-2):86-100.e15). The effects of the drug artemether were species-specific and cell-type-specific. In alpha cells, a fraction of cells increases insulin expression and gain aspects of beta cell identity, both in mouse and human samples. Importantly, researchers found that in human beta cells, there is no significant change in insulin expression, whereas in mouse islets, beta cells reduce their insulin expression and overall beta cell identity.
This study is the result of a cross-disciplinary collaboration of the laboratories of Stefan Kubicek and Christoph Bock at CeMM with Patrick Collombat at the Institute of Biology Valrose (France). This is the first study to apply single cell sequencing to analyze dynamic drug response in intact isolated tissue, which benefitted from the high quantitative accuracy of the decontamination method. It provides thus not only a novel method for single-cell decontamination and highly quantitative single-cell analysis of drug responses in intact tissues, but also addresses an important current question in islet cell biology and diabetes research. These findings could open up potential therapeutic avenues to treat Type 1 diabetes in the future.