Cell-free quest for new antibiotics
A new method combines synthetic biology with artificial intelligence
The rising resistance of bacteria to antibiotics presents an escalating global health risk. Now, researchers at the Max Planck Institute for Terrestrial Microbiology have combined synthetic biology and artificial intelligence (AI) to develop a more efficient approach to finding and creating new antimicrobial peptides that are effective against a wide range of bacteria.
Bioactive peptides play a key role in health and medicine. More than 80 peptide-based drugs are currently in use, all isolated from natural sources. However, antibiotic resistance is estimated to cause more than one million deaths worldwide each year. This number is expected to rise to 10 million by 2050, creating an urgent need for novel methods to accelerate the development of new antimicrobials. An untapped potential lies in the non-natural space, where an estimated number of 2010 to 2030 different peptides have yet to be explored.
In collaboration with several laboratories at the MPI for Terrestrial Microbiology, the University of Marburg, the MPI for Biophysics, the Bundeswehr Institute for Microbiology, the iLung Institute and INRAe France, a team of scientists at the Max Planck Institute led by Prof. Tobias Erb has established a new pipeline for the development of bioactive peptides.
"In deep learning, a neural network - algorithms inspired by the human brain - learns from large amounts of data. This type of machine learning holds great promise for peptide discovery and de novo design. However, it is usually followed by chemical synthesis of peptides for experimental validation, which is rather difficult and time-consuming and severely limits the number of peptides that can be chemically synthesized," explains Amir Pandi, lead author of the study.
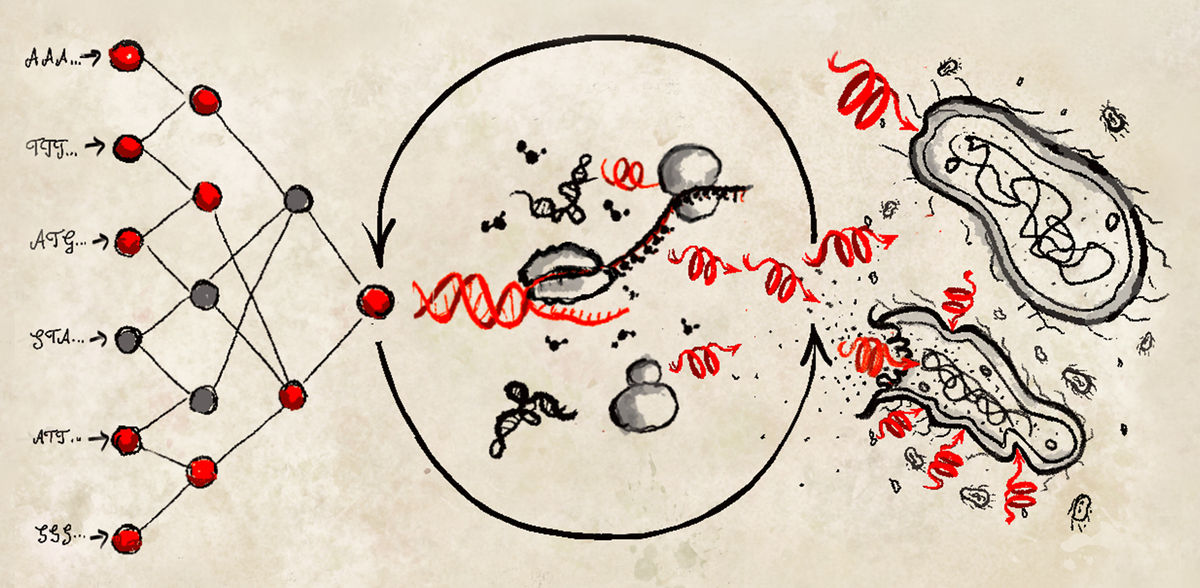
To overcome these limitations, the research team established a cell-free protein synthesis (CFPS) pipeline for the rapid and cost-effective production of antimicrobial peptides (AMPs) directly from DNA templates. The new protocol provides a rapid, low-cost, high-throughput method for AMP screening.
The team first used generative deep learning to design thousands of AMPs de novo, and then predictive deep learning to narrow them down to 500 candidates. Among these, screening with the cell-free pipeline identified 30 functional AMPs, which the researchers further characterized through molecular dynamics simulations, antimicrobial activity, and toxicity.
Notably, six of the de novo AMPs exhibited broad-spectrum activity against multidrug-resistant pathogens and did not develop bacterial resistance.
"We have benefited greatly from this combination of cell-free synthetic biology, artificial intelligence, and a high-throughput approach. By increasing the number of candidates that can be experimentally tested in less than 24 hours, the chance of finding active AMPs increased," says Amir Pandi. "Thus, our CFPS pipeline not only complements recent advances in computational design. It also has the potential to explore the relationship between design and function of bioactive peptides more quickly and cost-effectively." Tobias Erb adds: " This new method at the interface of synthetic biology and machine learning will be of interest to scientists working in the fields of biomedicine and bioactive peptides."
The next steps include further improving the yield of peptide production as well as employing AI and synthetic biology approaches to design new AMPs more stable, less toxic, or add a specific mode of action. The researchers also plan to apply augmented deep generative models where the machine learns molecular representations for desired properties, which would improve the success rate of identifying active candidates.
Original publication
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.