The enzyme with the iron heart
Results could lead to new biocatalysts for sustainable biotechnology
nitrogenases are the only enzymes capable of reducing atmospheric nitrogen to bioavailable nitrogen (ammonia), a process essential for all life on Earth. Using cryogenic electron microscopy, researchers at the Max Planck Institute for Terrestrial Microbiology in Marburg, Germany, have now solved the entire structure of the iron nitrogenase complex and revealed its unique molecular architecture. Their findings lay the groundwork for understanding the structure-function relationships of this important enzyme and could contribute to the development of novel biocatalysts for sustainable nitrogen assimilation and CO2 conversion.
Nitrogen is part of many important biomolecules, such as the bases of our DNA and the amino acids that make up proteins and enzymes. Before nitrogen can become part of biomolecules, it must be activated. In nature, only a single class of enzymes, called nitrogenases, can perform this life-sustaining catalysis.
Nitrogenases can also process CO2
Recently, it has been shown that nitrogenases can also reduce carbon dioxide (CO2) and carbon monoxide to hydrocarbons, like methane or ethylene, which in principle offers a way to recycle carbon waste into hydrocarbon products. Within the nitrogenase family, iron (Fe) nitrogenase is the isozyme with the highest natural CO2 reduction activity. However, the molecular architecture that enables these reactions has remained unknown, a problem that Dr. Johannes Rebelein's team at the Max Planck Institute for Terrestrial Microbiology set out to solve.
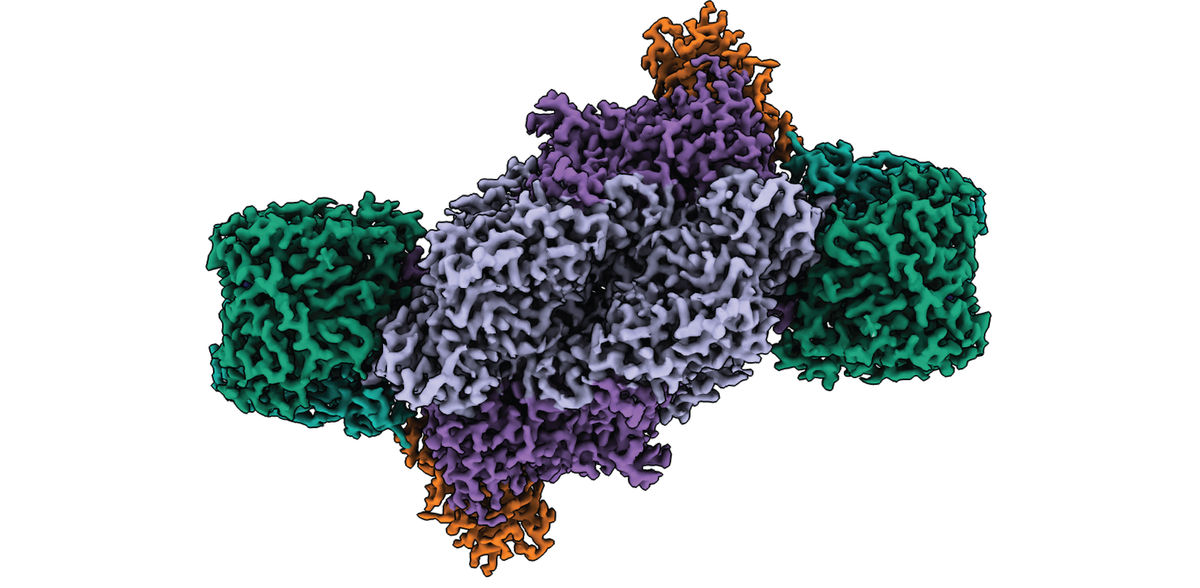
Using an engineered variant of the purple non-sulphur bacterium Rhodobacter capsulatus as their model organism, the researchers purified and biochemically characterized the Fe nitrogenase. By combining anaerobic biochemistry and cryogenic electron microscopy in collaboration with the Central Electron Microscopy Facility at the Max Planck Institute of Biophysics in Frankfurt, they have now solved the structure of the entire Fe Nitrogenase complex.
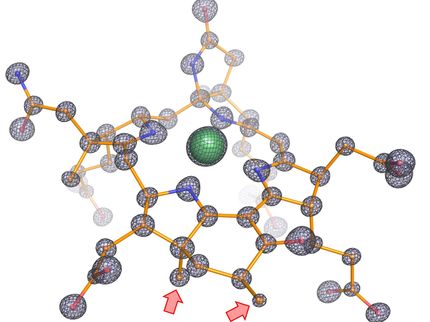
The results show that the active site cofactor in contrast to other types of nitrogenases, does not contain any metal other than iron. Moreover, the architecture of the Fe nitrogenase reveals distinct structural properties that may be responsible for the unique catalytic properties of the Fe nitrogenase.
Differences going beyond the active center
"We had expected that the main difference between Fe nitrogenase and other forms of nitrogenase, like the molybdenum nitrogenase, would be the architecture of the active site cofactor and its immediate environment. We were surprised to see that despite the differences in their catalytic properties, the active sites of the three nitrogenase isoforms are actually very similar,” says Frederik Schmidt, PhD student in the laboratory of Johannes Rebelein and co-first author of the study. But if there are no striking differences around the active site, which features might explain the unique reactivity of this enzyme?
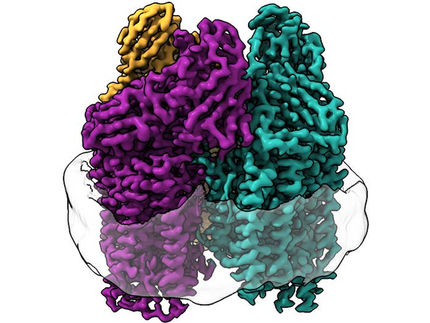
The researchers point to two structural features: First, the enzyme has an additional subunit called the G-subunit. Although this subunit has been described before, its function has remained elusive. Based on their new structure, the Max Planck researchers propose three possible functions for the G-subunit, including coordination of electron transfer, substrate channeling, and stabilization of the active site cofactor.
Increased reactivity through altered symmetry
Nitrogases are enzymes whose function is achieved through the interaction of two symmetrical halves. However, when the researchers superimposed the structure of Fe-nitrogenase on the already solved structure of Mo-nitrogenase, they found that the symmetry of the latter was different. "It is assumed that the two symmetrical halves communicate with each other in order to coordinate the catalytic mechanism of the enzyme. The altered symmetry that we observed in Fe nitrogenase could explain its particular reactivity," explains Luca Schulz, co-first author of the study.
How exactly these structural differences influence the catalytic mechanism of the Fe nitrogenase will be a key question for the future. Going forward, the researchers hope to further elucidate the molecular mechanism of the enzyme, which may help in the development of novel biocatalysts for sustainable nitrogen assimilation and CO2 conversion.