ONE microscopy: from molecule to 3D structure with conventional microscopy
The simple and cost-effective method developed using a few simple but effective tricks
Researchers at the University Medical Center Göttingen (UMG) have developed a new method that makes it possible for the first time to image the three-dimensional shape of proteins with a conventional microscope. Combined with artificial intelligence, One-step Nanoscale Expansion (ONE) microscopy enables the detection of structural changes in damaged or toxic proteins in human samples. Diseases such as Parkinson's disease, which are based on protein misfolding, could thus be detected and treated at an early stage. ONE microscopy was named one of the “seven technologies to watch in 2024” by the journal Nature and was recently published in the journal Nature Biotechnology.

The inventors (from left), group leader Dr. Ali H. Shaib and Professor Dr. Silvio O. Rizzoli, director of the Department for Neuro- and Sensory Physiology at the University Medical Center Göttingen (UMG).
A. Shaib
Fluorescence imaging is one of the most versatile and widely used tools in biology to observe biological processes in living cells. Despite advances in technology and improvements in resolution, the visualization of single molecules and the organization of molecular complexes using fluorescence microscopy remains a challenge. Until now, this was only possible using expensive structural biology methods such as electron microscopy (EM) and, in particular, cryo-EM, in which the samples are imaged with a very strong electron beam at an extremely low temperature.
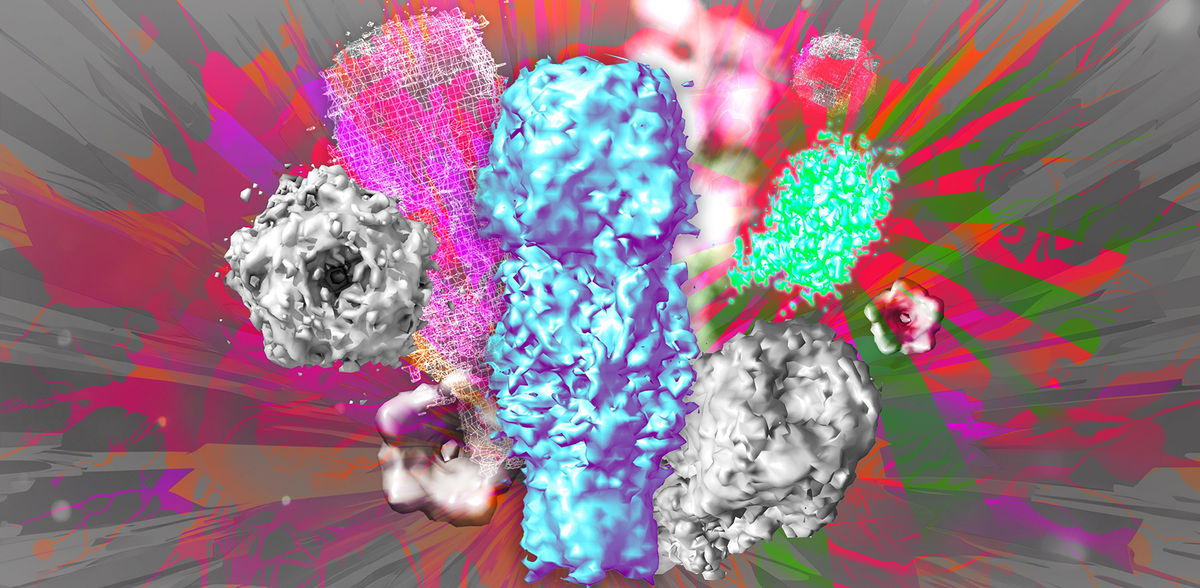
A research team led by Professor Dr. Silvio O. Rizzoli, director of the Department of Neuro- and Sensory Physiology at the University Medical Center Göttingen (UMG), spokesperson of the Center for Biostructural Imaging of Neurodegeneration (BIN) and member of the Cluster of Excellence “Multiscale Bioimaging: from molecular machines to networks of excitable cells” (MBExC), and Dr. Ali Shaib, group leader at the Department of Neuro- and Sensory Physiology at the UMG, have now developed a method using a few simple but effective tricks to visualize individual molecules in detail using conventional light microscopy. Instead of using expensive, high-resolution microscopes to improve the resolution, they developed One-step Nanoscale Expansion (ONE) microscopy. In this method, the volume of the sample is increased by binding the cells and the structures therein to a water-absorbing gel that penetrates the cells. By absorbing water, the gel increases up to 15 times its volume. This causes the molecules in the sample to move apart evenly and also become larger, so that they can be imaged with a light microscope after being specifically labeled with fluorescent molecules. Combined with a method based on artificial intelligence to evaluate the fluorescence changes, the scientists have succeeded for the first time in doing what was previously only possible with high-resolution cryo-electron microscopy and X-ray technology: “We are now able to reconstruct 3D protein structures from two-dimensional fluorescence images,” says Professor Rizzoli. This offers an unprecedented opportunity to directly visualize fine structural details of individual proteins as well as multiprotein complexes in cells or in isolation. Changes in the spatial structure of proteins can also be easily detected with ONE microscopy. In a collaboration with colleagues from Göttingen and Kassel, molecular protein aggregates, which are typical of Parkinson's disease, were imaged and classified in cerebrospinal fluid samples from patients. This is promising for improved early detection of Parkinson's disease, which affects millions of people worldwide.
ONE microscopy is a simple and cost-effective method that can be carried out in any laboratory with a conventional microscope and achieves a resolution level of around one nanometer. This is about 100,000 times smaller than the diameter of a human hair. The authors provide the necessary software as a free open source package. The newly developed method has now been published in the journal Nature Biotechnology. Due to its high potential, Nature has included ONE microscopy in its list of “seven technologies to keep an eye on in 2024”.
Original publication
Ali H. Shaib, Abed Alrahman Chouaib, Rajdeep Chowdhury, Jonas Altendorf, Daniel Mihaylov, Chi Zhang, Donatus Krah, Vanessa Imani, Russell K. W. Spencer, Svilen Veselinov Georgiev, Nikolaos Mougios, Mehar Monga, Sofiia Reshetniak, Tiago Mimoso, ... Julia Preobraschenski, Ute Becherer, Tobias Moser, Edward S. Boyden, A. Radu Aricescu, Markus Sauer, Felipe Opazo, Silvio O. Rizzoli; "One-step nanoscale expansion microscopy reveals individual protein shapes"; Nature Biotechnology, 2024-10-9