Rice University lab encodes collagen
Program defines stable sequences for synthesis, could help fight disease, design drugs
The human body is proficient at making collagen. And human laboratories are getting better at it all the time. In a development that could lead to better drug design and new treatments for disease, Rice University researchers have made a major step toward synthesizing custom collagen. Rice scientists who have learned how to make collagen – the fibrous protein that binds cells together into organs and tissues – are now digging into its molecular structure to see how it forms and interacts with biological systems.
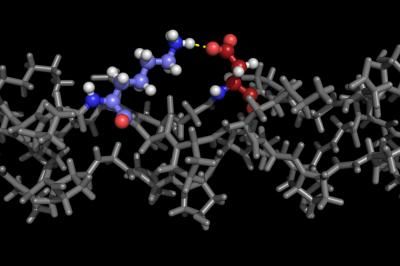
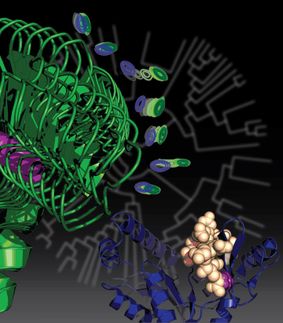
A program developed at Rice University details stable forms of collagen proteins for synthesis in the lab. The ability to synthesize custom collagen could lead to better drug design and treatment of disease. The colored portion of the molecule in this illustration shows positively charged lysine and negatively charged aspartate interacting in the required axial geometry that stabilizes the triple helix.
Hartgerink Lab/Rice University
Jeffrey Hartgerink, an associate professor of chemistry and of bioengineering, and his former graduate student Jorge Fallas, now a postdoctoral researcher at the University of Washington, wrote a new computer program that predicts the most stable structures of nanometer-sized collagen. In nature, these small structures link into chains that serve as connective tissue in the body. Hartgerink and Fallas followed up the computer research by making and testing the collagen detailed in their calculations.
Their success, reported in Nature Communications, will be of interest to physicians and scientists who work in reconstructive surgery, cosmetics and tissue engineering as well as to researchers investigating collagen protein interactions that could lead to new treatments for cancer and other diseases.
"Collagen is an odd protein. On one hand, it's the most abundant protein in the human body," said Hartgerink, who in a previous work unveiled a new way to synthesize self-assembling collagen. "It basically is the connective fiber that holds cells together; without it you'd turn into a big puddle.
"By mass, collagen is the most common protein there is. But it's different from almost any other you might look at," he said.
Hartgerink likened collagen to DNA with a structural twist, as it has not two but three intertwining peptide strands. "Watson and Crick, when they were first trying to understand DNA, figured out the underlying code for how all the base pairs fit together," he said. "Collagen is similar, except there are three strands. In this paper, we've started to crack the code of which amino acids go with what others to stabilize the structure."
While scientists have made a great deal of progress defining the structures of other proteins, "only a small group of us have been interested in collagen. And because of that, our understanding of it has lagged behind," he said.
In their new work, Hartgerink and Fallas analyzed charged interactions between amino acids that attract one strand to another (and in this case, yet another) to form the triple helix. "We look at positively charged and negatively charged amino acids and where they need to be aligned to result in stabilization," Hartgerink said.
In the same way three-color images must be properly aligned for a viewer to see a complete picture, the three strands of a collagen protein must be in register for the protein to carry out its function.
"Collagen does more than hold cells together," he said. "It also binds other proteins that have interesting functions. Those proteins will attach to collagen, and then cells come along and bind to those proteins. Based on that interaction, a cell will then 'decide' how to behave or differentiate into a different type of cell."
Hartgerink said that property makes collagen especially valuable for biological scaffolds, materials that are under intense study as a way to grow new body parts – even entire organs – to replace damaged ones.
Hartgerink said strand alignment also determines a collagen helix's stability. The computer program designed by Fallas and Hartgerink calculates the stability of each possible alignment of a given set of peptide strands – 27 in all – to find the best matches of positively and negatively charged amino acids. It then assigns each set a score, based on the net positive or negative charge of the entire helix.
"If we have a positive charge in a peptide sequence, it will destabilize the triple helix, and we score that as a minus 1," he said. "If we have a negative charge, that also destabilizes the helix and we also score that as a minus 1. But if those charges line up in what we call the axial geometry, it negates the destabilization. This triple helix would have a score of 0, which is good.
"We create huge, theoretical populations of collagen sequences and score all of them," he said. "We find out which are closest to this magical score of 0 and throw out all the other ones." That tells the researchers which sequences are likely to self-assemble into the most stable helixes. "The math looks complicated, but a personal computer can generate one of these sequences in one or two minutes of processing time. It's not super sophisticated." He said the code will be available on his group's home page for other researchers to try.