The Number One of Enterobacteria Deciphered
Genome of the Escherichia coli type strain finally sequenced — DNA includes potentially pathogenic segments
The colon bacillus Escherichia coli is one of the best studied model organisms in the life sciences. However, the reference organism for this species, its so-called type strain, has been overlooked in microbial genomics until now. In the “Genomic Encyclopedia of Bacteria and Archaea” (GEBA) project, the DNA of type strain DSM 30083T has now been sequenced and compared to that of close relatives of the strain. This study not only allows an entirely new view of the numerous E. coli strains that play relevant roles in medicine and biotechnology, including the EHEC pathogen and Shigella, but they also yielded a generally applicable method for determining the subspecies of any bacterial species. The research was conducted at the Leibniz Institute DSMZ – German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany, and at the Joint Genome Institute, Walnut Creek, CA, USA.
The colon bacterium Escherichia (E.) coli to microbiologists and biotechnologists is like a “pet bacterium” and looks back on an exciting history. Initially described as "Bacterium coli commune" by bacteriologist Theodor Escherich in 1886, its original isolate was lost at the beginning of the 1920s. It was not until 1941 that it was isolated again, this time by Fritz Kaufmann at the State Serum Institute in Copenhagen, Denmark, who also deposited it in in several collections of microbial strains and provided a scientific description. Today, E. coli is likely the best understood microorganism in the world and serves as an important indicator for the quality of drinking and recreational waters.
"It seems strange that the number one, the type strain of a bacterium that has entire scientific conferences dedicated to it as a model organism, had not been fully sequenced until now", said Christine Rohde, Head of the E. coli strain collection at DSMZ, Braunschweig, Germany. "Initially, scientists primarily sequenced the genomes of pathogenic strains of E. coli, or of genetically modified strains of biotechnological relevance. In addition, physicians and hygienists in their daily practice use serotypes that are quickly determined by antibody tests in order to differentiate between different strains of E. coli.”
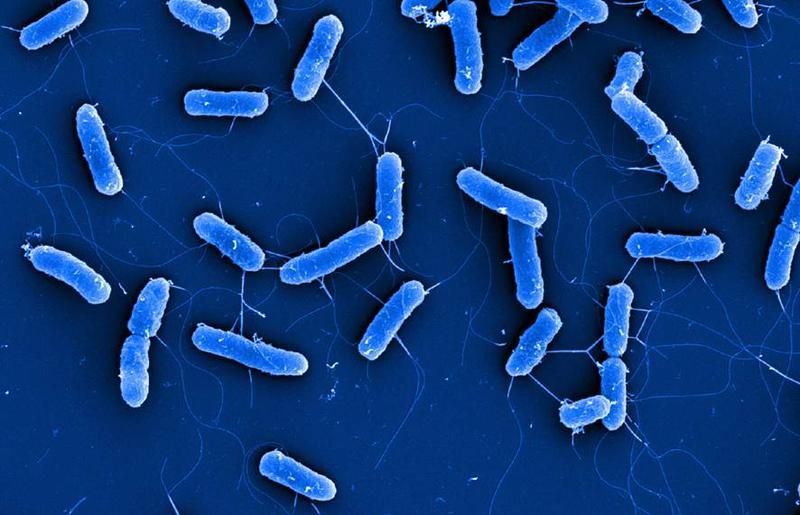
Scanning electron microscopic image of cells of E. coli type strain DSM 30083T
(courtesy of Manfred Rohde, Helmholtz Centre for Infection Research; Christine Rohde, Leibniz Institute DSMZ)
As Markus Göker, a bioinformatics scientist at DSMZ added: “Complete bacterial genomes are of fundamental importance for diagnostics in humans, for biotechnology, and for the search for antimicrobial agents. Today, this is truer than ever, as some strains of E. coli have developed into dangerous pathogens such as EHEC or EAHEC. The E. coli type strain was sequenced as part of the GEBA project that focuses on type strains exhibiting an unusual physiology or occupying a key place in the phylogenetic tree. This is the only microorganism in the project that was included based on its importance as a model organism.”
A genome with pathogenic potential
There are major physiological and genomic differences between the E. coli type strain and the harmless laboratory strain K-12. “Due to its serotype, the type strain had been grouped into the biological containment level 2, and its genome sequence now confirmed its pathogenic potential, “ said Jörn Petersen, an expert of plasmid biology at the DSMZ. “Unlike laboratory strain K-12, the E. coli type strain harbors an additional circular plasmid of 131,289 base pairs in its genome of 5,038,133 base pairs; this plasmid exhibits a sequence identity of 99% with plasmids from pathogenic E. coli isolates. These strains cause, e.g., colibacillosis in poultry and meningitis in newborns, with the horizontally transferable plasmid being responsible for their virulence,” explained Petersen.
Sophisticated computer-aided phylogenetic analysis
Thanks to the complete genome sequence of the E. coli type strain, the Braunschweig scientists were able to examine whether the huge number of previously sequenced isolates of E. coli actually belong to the same species, using modern taxonomic techniques in the process. “To this end, we analyzed more than 250 strains of E. coli and also verified their published taxonomic classification in subgroups, the 'phylotypes'. This bioinformatics-based analysis was performed with the state-of-the-art GGDC method. This technique is analogous to classical DNA-DNA hybridization in the laboratory, but yields significantly more exact results," as Markus Göker explained.
The analysis confirmed that all sequenced strains of E. coli belong to the same species. What is new, however, is the realization that E. coli is to be classified as having several subspecies. One of these subspecies includes all strains of the genus Shigella, known to cause shigellosis. “However, the name Shigella has historically been established in medicine, so we were not striving for taxonomic changes in this case,” Markus Göker added. “What is much more important is that the techniques tested in E. coli can now been used to classify bacterial species into subspecies in general.”
Original publication
Meier-Kolthoff JP et al. (2014); "Complete genome sequence of DSM 30083T, the type strain (U5/41T) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy."; Stand Genomic Sci 9: 2