HyPer-Tau provides spatially-resolved hydrogen peroxide sensing in cells
By attaching a hydrogen peroxide reporter protein to cellular microtubule structures, researchers have developed the first sensor able to show the location of the key cellular signaling chemical inside living cells with high resolution over time.
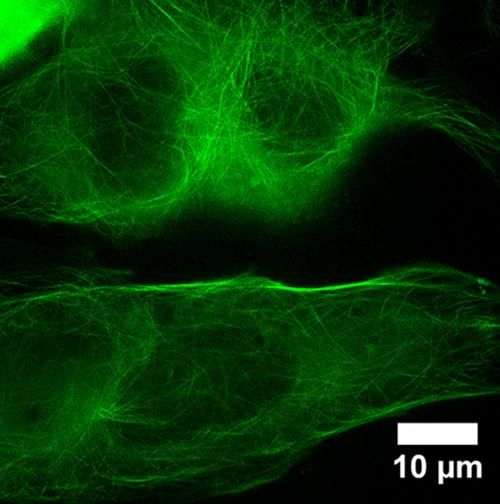
This super-resolution fluorescence microscopy image of HyPer-Tau shows the microtubular structure of a human cancer cell. The image was made using the new super-resolution microscope in the Georgia Tech Institute for Bioengineering & Bioscience.
Emilie Warren, Georgia Tech
Knowing the precise location of hydrogen peroxide within cells could help scientists gain a better understanding of oxidation-reduction reactions taking place there. The sensor was developed by researchers at the Georgia Institute of Technology, who have demonstrated several applications for its ability to spatially resolve the chemical's presence inside cells. Known as HyPer-Tau, the new sensor modifies a commercially-available protein that alters its fluorescence properties in the presence of hydrogen peroxide.
"The chemistry of cells, unlike more traditional chemistry in test tubes, is highly dependent on where a chemical reaction is occurring," said Christine Payne, an associate professor in the Georgia Tech School of Chemistry and Biochemistry and one of the paper's senior authors. "HyPer-Tau is a tool that will provide us with information on the 'where' and 'when' for hydrogen peroxide inside living cells."
Until development of the new technique, hydrogen peroxide sensors could only tag certain components of cells, or show that the cells contained the oxidant. To understand the role of hydrogen peroxide in signaling and oxidation, however, the researchers wanted to know the time-resolved location of the chemical.
"We needed a tool that could discriminate between locations to provide more than a whole readout of oxidation," said Melissa Kemp, an associate professor in the Wallace H. Coulter Department of Biomedical Engineering at Georgia Tech and Emory University. "With very specific spatial information, we could be better informed about how cellular processes or therapies involving oxidation are going to operate."
Kemp and Payne realized that if they could anchor the reporter protein to microtubules - fibrous structures that crisscross cells like railroad tracks - they might obtain the location information they needed.
Other researchers had already created variants of the HyPer reporter protein, so the researchers - with technician Emilie Warren, undergraduate researcher Tatiana Netterfield and postdoctoral researcher Saheli Sarkar - set out to create a new tool. They added a tubulin-binding protein known as Tau, that connects the HyPer protein to the microtubule structures. Fluorescence microscopy then allowed them to observe the real-time change in fluorescence as oxidation occurred in the cells they were studying.
"Connecting the reporter protein allows us to get a grid-type readout of oxidation going on inside the cells," said Kemp. "By having the protein tethered, we can get very specific sub-cellular information. You can readily see areas with more intense oxidation."
She used a traffic analogy to compare information provided by the new technique to that provided by the earlier one. Earlier sensors would have reported that traffic in a downtown area was congested, while the new sensor could pinpoint an accident on a specific street causing the delays. The latter information allows specific action to be taken, Kemp said.
Kemp and Payne have already used the tool to visualize the signaling process that takes place as macrophages discover bacteria and move to engulf and destroy the invaders.
"When the macrophages are activated, they begin shooting out tiny leg-like structures that seek the bacterial signal," explained Kemp. "To do so, they require hydrogen peroxide to control the migration and other activities. We can see in these leading edges where the oxidation is occurring inside the cells, providing an unprecedented view of the behavior."
By combining multiple images, the researchers produced movies correlating the production of hydrogen peroxide to the activities of the immune system cells.
In another application, the sensor was used to study how cells respond to the introduction of extracellular hydrogen peroxide, which produces a wave of oxidation as it moves through the cellular structures.
"This provides a way to quantify both intracellular and intercellular variation that is occurring," Kemp explained. "Our goal is to be able to monitor in real-time the events that are occurring. Because of the spectral features of the reporter, you can couple this with other types of dyes to monitor organelles and different types of production."
Kemp hopes to use the new sensor to better understand oxidation of another type of immune cell, T cells, as they form contact with other cells to recognize the presence of viruses. In studies that could be important to understanding the effects of nanoscale materials on living cells, the researchers are working to understand the suspected oxidative impacts of titanium dioxide nanoparticles. The new technique could also be useful in understanding how stem cells change oxidation properties during differentiation into other cell types.
In current research, Netterfield is working with Kemp and Payne to combine the existing technique with other reporter proteins to gain additional information.
Once thought to be a sign of disease processes, hydrogen peroxide is now understood to be a critical signaling chemical inside cells, Kemp noted. Cells purposely produce the chemical, which can quickly oxidize proteins to alter their functions. Hydrogen peroxide is also generated at sites of inflammation, and as macrophages destroy pathogens.
Collaboration between Payne - a physical chemist - and Kemp - a biomedical engineer, demonstrates how innovation can occur at the intersections of disciplines.
"Chemistry and biomedical engineering offer a pretty natural collaboration," said Payne. "We both speak the same science language and have a shared interest in developing new tools to enable new science."