Neutrons help to determine enzyme function
New point of attack against stomach bacterium Helicobacter pylori
There is a strong suspicion that Helicobacter pylori is linked to the development of stomach cancer. Now an international team of researchers led by Prof. Donald R. Ronning (University of Toledo, USA) has used neutrons to unlock the secret to the functionality of an important enzyme in the bacterium's metabolism. This could be used as a point of attack for new medications. The team made the corresponding measurements at the neutron sources in Oak Ridge (USA) and at the research neutron source reactor FRM II of the Technical University of Munich (TUM).
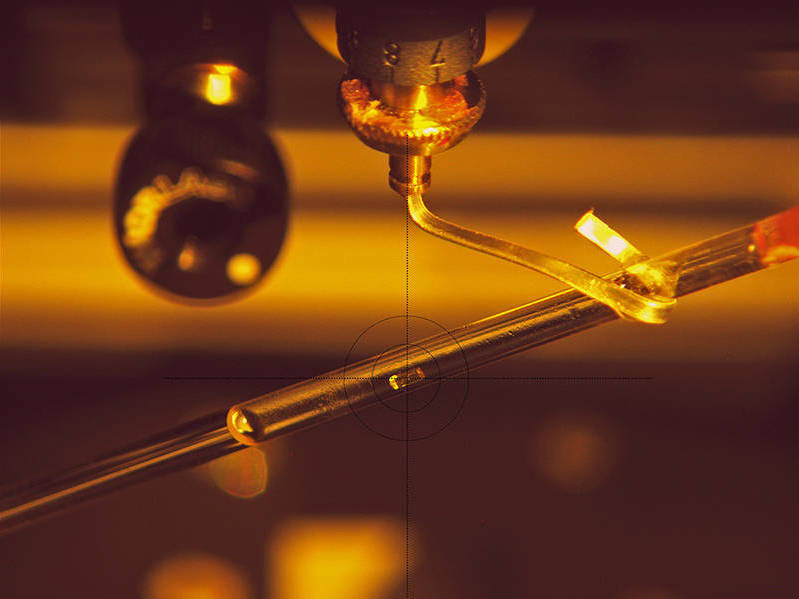
Placing a crystal of the enzyme MTAN in the center of the neutron beam at the instrument BIODIFF
Andreas Ostermann / TUM
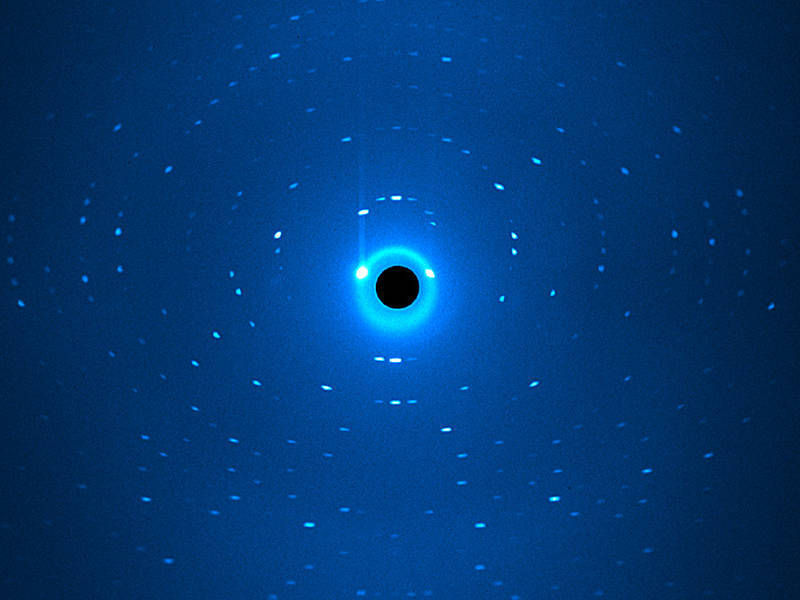
Diffraction image resulting from neutron scattering at the instrument BIODIFF
Andreas Ostermann / TUM


One in every two people everywhere in the world has it in his or her stomach: Helicobacter pylori. Ulcers and chronic gastritis are the most common illnesses associated with this bacterium. Up to now the standard therapy used to combat this stomach germ has been a combination of two antibiotics and a proton pump inhibitor. But this treatment is only successful in 70 percent of cases, and an increasing level of resistance is developing. For quite some time now scientists have therefore been searching for alternative medications to fight this dangerous bacterium.
In contrast to humans and to many helpful bacteria, H. pylori uses a special enzyme to synthesize vitamin K2. As a result, this enzyme, 5'-methylthioadenosine nucleosidase (MTAN), offers very promising prospects for the development of a medication that acts specifically against H. pylori without harming useful bacteria or even human cells.
Structural determination with neutrons
The enzyme MTAN is part of an important step in the synthesis of vitamin K2. Hydrogen bonds bind a precursor of the vitamin in order to cut off a side chain. But the positions and position changes of the hydrogen atoms essential to this process were not previously known with certainty.
The usual method of structural determination for enzymes, crystal structure analysis using x-ray radiation, is of little help here, since the x-ray radiation is almost blind to hydrogen atoms. Therefore the researchers based their structural determination on neutrons, which are particularly sensitive to hydrogen atoms.
The scientists investigated different variations of the enzyme at the BioDiff diffractometer, operated jointly by the TUM and the Jülich Centre for Neutron Science (JCNS) in the Heinz Maier-Leibnitz Zentrum in Garching north of Munich, and at the Oak Ridge National Laboratory (USA) neutron source. The joint measurements allowed them draw a detailed picture of the enzyme's mode of action.
"Now that we know the exact process of the reaction and the binding sites of the enzyme involved, it's possible to develop molecules that block exactly this process," says TUM biologist Andreas Ostermann, who supervises the instrument at the FRM II together with Dr. Tobias Schrader (JCNS).
The publication included researchers from the University of Toledo (Toledo, USA), Victoria University of Wellington (New Zealand), the Heinz Maier-Leibnitz research neutron source at the Technical University of Munich (TUM), the Jülich Centre for Neutron Science and the Oak Ridge National Laboratory (USA). The research was supported by funds from the Center for the Advancement of Science in Space (USA) and the National Institute of Allergy and Infectious Disease (USA). X-ray structure data were measured at the Advanced Photon Source of the Argonne National Laboratory (USA) with the support of the Michigan Economic Development Corporation.