Unmasking blood doping in athletes
RNA changes mark old blood cells among new ones, foiling self-transfusions
A Duke University research team has found a way to help sporting officials detect whether an athlete's blood has been doped by an infusion of their own stored blood.
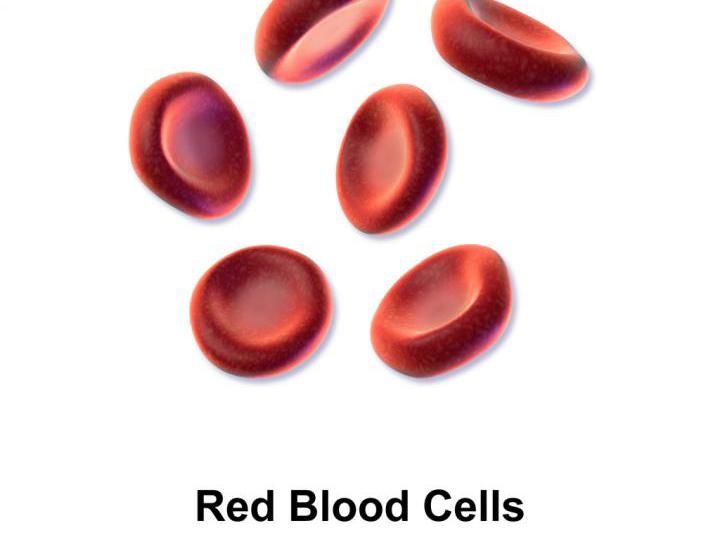
Red blood cells ferry oxygen to tissues, providing fuel for muscular activity. Unscrupulous athletes have been able to 'dope' their blood by enriching their supply of red blood cells. But a new test of microRNA can tell old cells from new ones.
Bruce Blausen via Wikimedia Commons
While tests have been developed to detect two of the three most common methods of dramatically boosting the oxygen-carrying capacity of a competitor's blood, so-called "autologous" or self-transfusions have been impossible to detect.
An autologous transfusion takes some of the athlete's blood out well before the competition, sorts out just the red blood cells, and then transfuses those cells back into the athlete right before competition to enhance the blood's ability to carry oxygen, the essential fuel of muscle performance.
The best detection method the World Anti-Doping Agency (WADA) has used to date is the "Athlete Passport," which compares a pre-competition blood sample to one taken at competition to see if there are 'significant' changes in biochemistry.
"The difficulty has been that the tests they have couldn't tell the difference between a young blood cell and an old one," said Jen-Tsan "Ashley" Chi, M.D., Ph.D., who led this WADA-funded research in his lab at Duke's Center for Genomic and Computational Biology.
Blood banks in the U.S. consider 42 days the outer limit of how long a unit of red blood cells should be stored because of biochemical changes that could harm recipients. The amount of energy-providing ATP drops and oxygen-binding hemoglobin declines as well. But those changes have not been precise enough to detect an autologous transfusion.
What Chi and his colleagues looked at in the red blood cells is nucleic acids, specifically RNA. Red blood cells were long thought to lack nucleic acids because they don't carry a nucleus, where one would normally find DNA. But it turns out they contain an abundant and diverse population of RNAs. Among these are some short RNA pieces called microRNAs (miRNA) which generally act to control the production of proteins in a cell.
The researchers drew three units of blood from volunteers and processed them to remove virtually all of the white blood cells and about 80 percent of the plasma, leaving behind a relatively purified sample of red blood cells, just as an autologous transfusion would require.
Then Jennifer Doss, a former Duke graduate student, and other lab members extracted and analyzed RNA samples taken from the cells at eight time intervals, from 1 day to 42 days. Changes in the RNA associated with storage became apparent as they compared the later samples to the Day 1 sample.
Two types of miRNA increased in number during storage and two declined, said graduate student Wen-Hsuan Yang, who performed the biochemistry experiments. One of the forms that declined, called miR-70, had the most dramatic and consistent changes.
With further testing, the researchers isolated the likely source of this 18-nucleotide fragment of RNA. It seems to result as a byproduct from a larger RNA that is cut by enzymes during storage, and it happens in a very precise, predictable way.
"This increase in miR-720 is significant enough and consistent enough that it could be used as a biomarker for detecting stored red blood cells," Chi said.
He said further research is focused on understanding why the enzyme that produces miR-720 is active in stored cells and what it might be doing as it breaks a larger RNA apart.
Original publication
Wen‐Hsuan Yang, Jennifer F. Doss, Katelyn A. Walzer, Shannon M. McNulty, Jianli Wu, John D. Roback, Jen‐Tsan Chi; "Angiogenin‐mediated tRNA cleavage as a novel feature of stored red blood cells"; British J Hematology; 2018