Structure of tuberculosis drug target determined
Rutgers University scientists have determined the three-dimensional structure of the target of the first-line anti-tuberculosis drug rifampin. They have also discovered a new class of potential anti-tuberculosis drugs that kill rifampin-resistant and multi-drug-resistant tuberculosis bacteria.
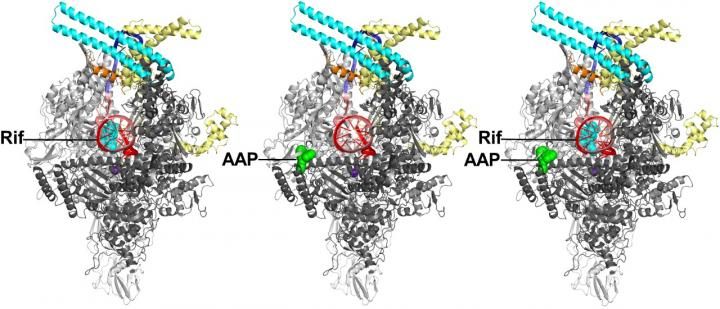
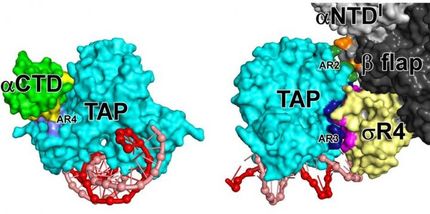
Crystal structures of Mtb RNAP bound to rifampin (left), Mtb RNAP bound to an AAP (center), and Mtb RNAP bound to both rifampin and an AAP (right) show that rifampin and the AAP interact with non-overlapping binding sites on Mtb RNAP and, as a result, can bind simultaneously to Mtb RNAP. Gray and cyan ribbons, Mtb RNAP; yellow and orange ribbons, transcription initiation factor sigma; red and pink ribbons, DNA (specific sequence elements in blue).
Wei Lin and Richard H. Ebright
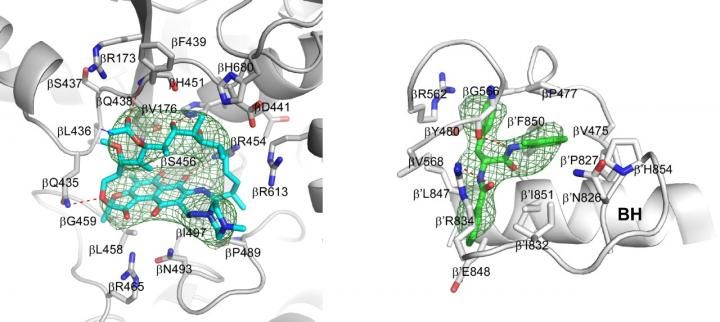
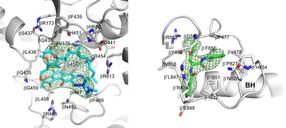
Crystal structures of Mtb RNAP bound to rifampin (left) and Mtb RNAP bound to an AAP (right) show the Mtb RNAP interactions. Gray ribbons, Mtb RNAP backbone; gray, cyan, and green sticks, Mtb RNAP, rifampin, and AAP carbon atoms; red and blue sticks, oxygen and nitrogen atoms; green mesh, mFo-Fc electron density omit map; BH, Mtb RNAP bridge helix.
Wei Lin and Richard H. Ebright
Tuberculosis (TB) bacteria infect a third of the world's population and the disease kills 1.8 million people annually.
Rifampin, a compound that kills TB bacteria, has been the cornerstone of anti-TB therapy since its discovery in 1961. However, rifampin-resistant TB has spread widely, posing an urgent public health crisis. Rifampin resistant TB arises when TB bacteria acquire mutations that alter the binding site for rifampin on the enzyme it inhibits in TB bacteria, Mtb RNA polymerase (Mtb RNAP). The alterations of the rifampin binding site prevent rifampin from binding to and inhibiting Mtb RNAP, preventing rifampin from killing TB bacteria.
Intensive efforts have been underway worldwide to develop improved rifampin derivatives that are unaffected by alterations of the rifampin binding site on Mtb RNAP and that thus can kill rifampin-resistant TB bacteria. Intensive efforts also have been underway to develop new, non rifampin-related Mtb RNAP inhibitors that function through binding sites on Mtb RNAP that do not overlap the rifampin binding site and that thus can kill rifampin-resistant TB bacteria. However, until now, these efforts have been hampered by the absence of structural information for Mtb RNAP, making rational, structure-based drug discovery for Mtb RNAP impossible.
Richard H. Ebright and other Rutgers scientists report the three-dimensional structures of Mtb RNAP and Mtb RNAP bound to rifampin. The structures were determined by use of X-ray crystallography and are at a resolution sufficient to define the positions, conformations and interactions of individual amino acid residues of Mtb RNAP. The results reveal the interactions between Mtb RNAP and rifampin, reveal the mechanism by which rifampin inhibits Mtb RNAP, and enable rational, structure-based design of improved rifampin derivatives for inhibition of Mtb RNAP.
In addition, the Rutgers scientists report the discovery and properties of new, non‑rifampin‑related compounds--Na-aroyl-N-aryl-phenylalaninamides (AAPs)--that potently and selectively inhibit Mtb RNAP and potently and selectively kill TB bacteria.They also report the structures of Mtb RNAP bound to an AAP and Mtb RNAP bound to both an AAP and rifampin. The results show that AAPs inhibit Mtb RNAP through a binding site that does not overlap the rifampin binding site and thus can inhibit rifampin-resistant Mtb RNAP and kill rifampin-resistant TB bacteria. The results further show that AAPs function additively if co-administered with rifampin and show that AAPs suppress the emergence of resistance if co-administered with rifampin. Taken together, the results show that AAPs are exceptionally promising new lead compounds for anti-TB drug development.
"The structure of Mtb RNAP has been the 'Holy Grail' for TB drug discovery targeting Mtb RNAP," said Ebright, Board of Governors Professor of Chemistry and Chemical Biology and Laboratory Director at the Waksman Institute of Microbiology at Rutgers.
"AAPs represent an entirely new class of Mtb RNAP inhibitors and are, without question, the most promising Mtb RNAP inhibitors for anti-TB drug development since rifampin," Ebright said. "We are very actively pursing AAPs. We have synthesized and evaluated more than 600 novel AAPs and have identified AAPs with high potencies and favorable intravenous and oral pharmacokinetics."
"Tuberculosis (TB) remains a major global health problem and identifying new drug targets and candidate drugs is a major priority," said David S. Perlin, Executive Director and Professor at the Public Health Research Institute and Rutgers Regional Biocontainment Laboratory at the International Center for Public Health at Rutgers New Jersey Medical School in Newark.
"This work by the Ebright laboratory represents a major step forward by providing a detailed structural look at RNA polymerase, an essential enzyme and a well-established drug target in Mycobacterium tuberculosis," Perlin said. "Furthermore, they demonstrate that a new chemical class binds to an alternative site on the enzyme, which overcomes existing drug resistance and opens up the exciting possibility for development of a novel class of anti-TB drugs."
Original publication
Wei Lin and Soma Mandal and David Degen and Yu Liu and Yon W. Ebright and Shengjian Li and Yu Feng and Yu Zhang and Sukhendu Mandal and Yi Jiang and Shuang Liu and Matthew Gigliotti and Meliza Talaue and Nancy Connell and Kalyan Das and Eddy Arnold and Richard H. Ebright; "Structural Basis of Mycobacterium tuberculosis Transcription and Transcription Inhibition"; Molecular Cell; 2017