New 3-D imaging reveals how human cell nucleus organizes DNA and chromatin of its genome
Sixty-four years ago, James Watson and Francis Crick described the now-iconic double helix structure of DNA. A team of researchers at the University of California San Diego School of Medicine and the Salk Institute for Biological Studies describe development and application of new electron microscopic imaging tools and a selective stain for DNA to visualize the three-dimensional structure of chromatin -- a complex of molecules that helps pack six feet of DNA into each cell nucleus, construct chromosomes and control gene expression and DNA replication.
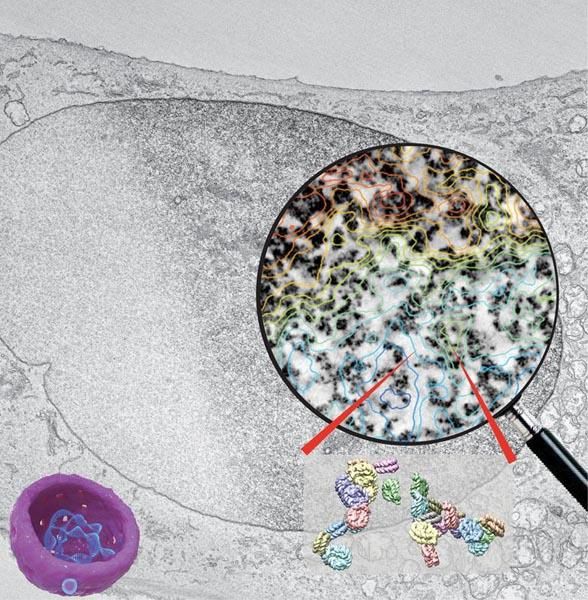
Chromatin structure determines the function of the human genome. To visualize chromatin of intact cells, a method called ChromEMT labels DNA and paints a metal dust over chromatin. In the composite image above, a black-and-white electron micrograph of a cell nucleus is half stained by ChromEMT. The stained nucleus is then imaged by electron microscopy to generate a 3D volume that reveals chromatin structure and organization. The circular magnified image shows ChromEMT stained chromatin (dark structures) overlapping with a contour map of chromatin density (blue-low, green-medium, red-high density). Analysis reveals that chromatin is organized as 5 to 24 nm diameter polymer chains that pack in wide distribution of density in interphase resting cells, but uniform density in mitotic cells. Two models of nucleosome orientations in the chromatin chain are shown at bottom right.
Horng D. Ou, Salk Institute.
"The primary functions of chromatin are fundamental," said study co-lead Mark Ellisman, PhD, Distinguished Professor of Neurosciences and Bioengineering and director of the National Center for Microscopy and Imaging Research (NCMIR) at UC San Diego. "It efficiently packages DNA to fit inside the cell nucleus, making it possible for chromosomes and cells to divide and replicate safely and correctly. It's a basic, working element of life."
The new findings and the methods developed allow them help resolve an on-going, persistent debate about the actual structure of chromatin, long poorly understood. DNA wraps around complex structures called nucleosomes. This chain of disks was thought to organize into increasingly thicker fibers that progressively form what are seen as condensed chromosomes in dividing cells.
Ellisman, with co-senior author Clodagh O'Shea, PhD, an associate professor at Salk and Howard Hughes Medical Institute Faculty Scholar, and colleagues demonstrate that this model of hierarchical packing into thicker and thicker cables is not correct. They developed a new imaging approach called ChromEMT, which combines an advanced form of electron microscopy tomography, developed at NCMIR, with a new labeling method to selectively enhance electron scattering and thus the specific contrast associated with DNA to directly image within cells the threads of this important core element of the genome.
Ellisman said that it is now clear that the biological functions and activity of our genomes in the nucleus are not determined by linear DNA sequence information alone. Instead, it is the local nucleosome structure combined with its global 3D organization in the nucleus that determines gene expression and cell fate. ChromEMT enables DNA and chromatin to be visualized across this critical set of biological and structural scales in single cells for the first time.
O'Shea said the findings reveal that nucleus DNA assembles five to 24-nanometer-diameter chromatin chains in a diversity of 3D conformations and motifs. "In contrast to ordered and rigid fibers, chromatin is a flexible chain that can collapse and pack together into 3D domains that have a wide range of different concentration densities," she said. "This provides exciting new insights into how different gene sequences, interactions and epigenetic modifications can be integrated at the level of chromatin structure to regulate gene expression and inherited and maintained through cell division."
O'Shea and Ellisman are co-principal investigators in a project funded by the 4D Nucleome Consortium to better understand how DNA is arranged within a cell nucleus in three dimensions plus time; and how these changes in DNA organization affect human health and disease. Their goal is to use ChromEMT to "crack the cell nucleus" and decipher how a cell's nucleus or control center oversees cell growth, metabolism and reproduction. They are now developing additional labels that can be combined with ChromEMT to visualize the structural basis of gene silencing and how viral and cancer proteins remodel DNA to drive pathological replication.