Case Western Reserve University researcher improves LCDs with 3-D nanoimaging process
Charles Rosenblatt, professor of physics and macromolecular science at Case Western Reserve University, and his research group have developed a method of 3D optical imaging of anisotropic fluids such as liquid crystals, with volumetric resolution one thousand times smaller than existing techniques. A research paper detailing the team's findings appeared in the advanced online publication of Nature Physics.
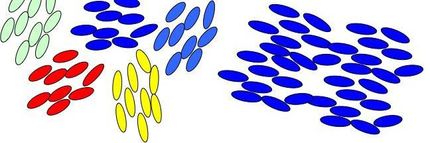
The molecules of these fluids, such as liquid crystals and ordered polymers, gels, and emulsion, can be oriented by magnetic or electric fields and thus can control the polarization properties of light. This is how liquid crystal displays in televisions, laptop computers, and other digital devices operate. Designing these and future devices requires a detailed knowledge of the molecular order. Until now much of the available information was based on inference from macroscopic experiments.
Using Rosenblatt's new technique, which provides detailed visual renderings of structures at the level of tens of nanometers, about 1/1000th the diameter of a human hair, we are able to create a much more detailed and nuanced picture of the structure. This will facilitate improvements to existing devices and make entirely new applications possible. Moreover, many fundamental scientific questions that deal with phase transitions or the nature of topological defects can be studied in far more detail than previously possible.
His system builds on existing techniques in near field scanning optical microscopy (NSOM) for 2D imaging. Traditionally, NSOM entails placing a very tiny optical fiber close to the specimen surface, in this case the substrate on which the anisotropic fluid resides. As the substrate is scanned back and forth below the fiber's aperture, a computer records the intensity of the light that emerges from the fiber and interacts with the surface, as well as the position of the fiber. The computer processes the information and produces a 2D optical image of the surface with resolution of several tens of nanometers. To date, NSOM has been used mainly as a surface characterization technique.
Rosenblatt's team adapted traditional NSOM technology by using polarized light, immersing the fiber into the fluid, and collecting images at a series of heights above the substrate. The result is polarized optical nanotomograpy (ONT), a system for 3D mapping of anisotropic fluid on top of a substrate.
The team chose a nematic liquid crystal, whose molecular orientation is controlled by a nanoscopic pattern scribed into the underlying polymer-coated substrate. This material was chosen because its structure can be readily calculated, giving the team an idea of what they should expect to see through the ONT imaging process.
In their ONT experiment the team immersed an optical fiber with a diameter of 60 nanometers (approximately one-tenth the diameter of the wavelength of light) into the liquid crystal to a position just above the substrate. There the fiber was scanned two-dimensionally over the surface and an image was obtained. They then retracted the fiber by approximately 25 nm and obtained a second image. In principle data from the first image could be subtracted from the second, providing information about the molecular orientation profile at 25 nm above the surface. This process was repeated out to a distance of 500 nm above the substrate. Rosenblatt's team demonstrated that the images at each height were completely consistent with the theoretical predictions, and they were able to make the first visualization and direct measurement of the 200 nanometer length over which the molecular orientation homogenizes.
Most read news
Organizations
These products might interest you

CHSN-O, CN and N Elemental Analyzers by Velp Scientifica
State-of-the-art Elemental Analyzers for N, CN and CHSN-O in organic samples
Consistency, ease of use, and premium features for elemental analysis following official standards

HYPERION II by Bruker
FT-IR and IR laser imaging (QCL) microscope for research and development
Analyze macroscopic samples with microscopic resolution (5 µm) in seconds

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.