Technology measures tumors' drug resistance up to 10 times faster
A group of scientists from VCU Massey cancer Center and UCLA Jonsson Comprehensive Cancer Center have developed a new, high-speed microscopy platform that can measure a cancer cell's resistance to drugs up to 10 times faster than existing technology, potentially informing more effective treatment selection for cancer patients.

This is an HLSCI image of live melanoma cells colored to indicate the distribution of mass within each cell.
VCU Massey Cancer Center
Fast and repeated assessments of tumor sensitivity to available drugs can improve clinical outcomes by staying ahead of a cancer's ability to resist certain therapies. Quantitative phase imaging has proven to be effective in measuring individual cell growth in response to various therapies, especially in cancers that grow in clusters such as melanoma. However, in current implementations, this technique is constrained by limited sample size.
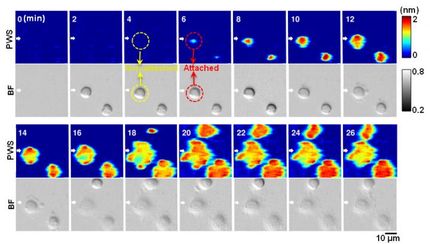
The group of scientists led by Jason Reed, Ph.D., member of the Cancer Molecular Genetics research program at VCU Massey Cancer Center, created a novel multi-sample, multidrug testing platform called High-Speed Live-Cell Interferometry (HSLCI) that directly measures tumor response to therapy at a significantly accelerated rate. HSLCI closely monitors the growth of a single cell's mass over time, but can measure the individual growth of 1,000 to 10,000 cells in one experiment, compared to previous studies which were limited to the analysis of approximately 60 cells per hour. If HSLCI measures a decrease in the growth of cancer cells, this is a sign that a treatment is effective.
"Our platform can detect effective versus ineffective drugs in patient-derived cells in a matter of hours, more than 10 times faster than the current standard. This speed and accuracy make HSLCI the first implementation of quantitative phase microscopy with the throughput required for a tool to guide patient therapy selection," said Kevin Leslie, M.S., a lead author of the study and PhD candidate in VCU's Integrative Life Sciences program. Leslie added that HSLCI can track cell behavior for more than five days, compared to a maximum duration of 48 hours common to most previous research.
In a study Reed applied HSLCI to models of metastatic melanoma, a disease in which tumor diversity and drug resistance are substantial obstacles to improved patient outcomes. Up to 80 percent of advanced melanoma patients receiving combination therapy develop drug resistance upon treatment.
There are presently no reliable cellular biomarkers that can guide targeted melanoma therapy or improve patient outcome predictions, and measuring drug response in vitro (in cells removed from the organism) often requires long turnaround times.
"A superior, rapid, accurate and inexpensive approach to determine melanoma drug sensitivity before - and periodically during - therapy is desirable," said Reed, an associate professor of physics at Virginia Commonwealth University.
Using industrial-grade imaging hardware, low-cost, multi-core PC processors and additional software improvements, Reed designed a completely new platform to combat these inefficiencies. The HSLCI takes images of standard-format, multi-well cell culture plates where each well can hold a different cell type exposed to a unique drug dose or combination. Requiring no fluorescence or dye labeling, it is well-equipped for noninvasive measurements of single cells and cell clusters.
Reed's research found that HSLCI successfully differentiated between thousands of drug-sensitive and drug-resistant melanoma cells in 24 hours.
"HSLCI could be used throughout all forms of cancer therapy to evaluate the efficacy of treatment. This assessment could ensure all cancer patients are receiving the most effective drugs for their disease," Reed said.
Reed hopes he can further develop this technology in preclinical animal tests and early-phase clinical trials, using breast cancer models in collaboration with Chuck Harrell, Ph.D., a member of Massey's Cancer Molecular Genetics research program.
Original publication
Dian Huang, Kevin A. Leslie, Daniel Guest, Olga Yeshcheulova, Irena J. Roy, Marco Piva, Gatien Moriceau, Thomas A. Zangle, Roger S. Lo, Michael A. Teitell, and Jason Reed; "High-Speed Live-Cell Interferometry: A New Method for Quantifying Tumor Drug Resistance and Heterogeneity"; Analytical Chemistry; 2018