Stone age hepatitis B virus decoded
Viral DNA shows the hepatitis B virus has been circulating in Europe for at least 7.000 years
An international team of scientists led by researchers at Kiel University in the scope the CRC 1266 “Scales of Transformation” and the Max Planck Institute for the Science of Human History has successfully reconstructed genomes from Stone Age and Medieval European strains of the hepatitis B virus. This unprecedented recovery of ancient virus DNA indicates that hepatitis B was circulating in Europe at least 7.000 years ago. While the ancient virus is similar to its modern counterparts, the strains represent a distinct lineage that has likely gone extinct.
The hepatitis B virus (HBV) is one of the most widespread human pathogens known today, affecting over 250 million people worldwide. However, its origin and evolutionary history remain unclear. Studying the evolution and history of the virus has to date been especially difficult, because until now viral DNA had not been successfully recovered from ancient samples. In the present study, published in eLIFE, an international team of researchers led by the Max Planck Institute for the Science of Human History and the Institute of Clinical molecular biology at Kiel University (IKMB), not only recovered ancient viral DNA but also reconstructed the genomes of three strains of HBV, two from the Neolithic and one from the medieval period.
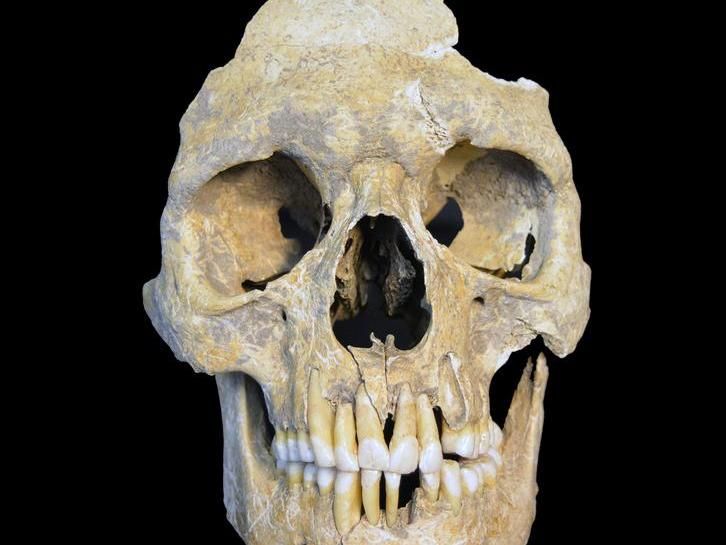
Skeletal remains of HBV positive individual from the Stone Age site of Karsdorf, Germany, it is from a male with an age at death of around 25-30 years.
Nicole Nicklisch
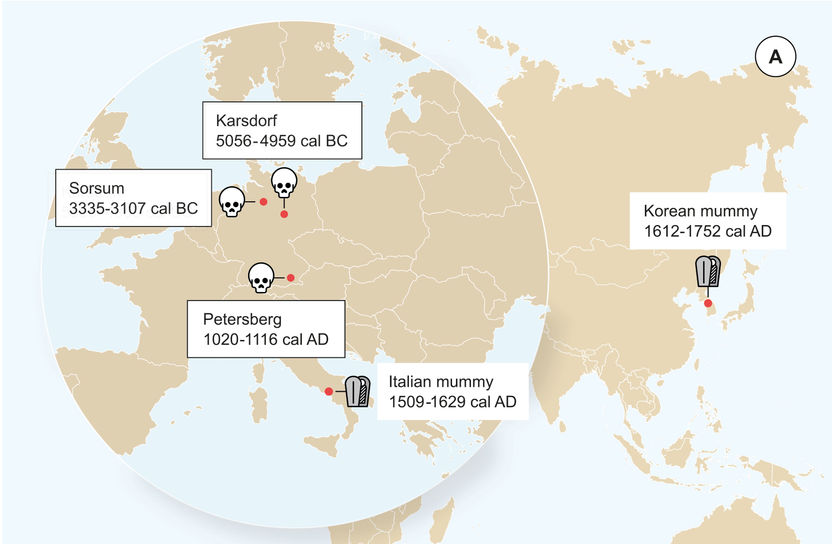
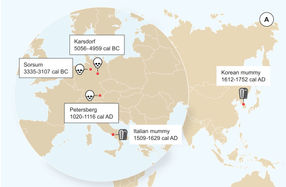
Geographic location of the samples from which ancient HBV genomes were recovered. Icons indicate the sample material (tooth or mummy). HBV genomes obtained in this study are indicated by black frame.
Krause-Kyora et al. Neolithic and Medieval virus genomes reveal complex evolution of Hepatitis B. 2018.
The ancient and complex history of hepatitis B
For this study, the researchers analyzed samples from the teeth of 53 skeletons excavated from Neolithic and medieval sites in Germany. The remains dated from around 5.000 BC to 1.200 AD. The researchers screened all samples for viral pathogens and detected ancient HBV in three of the individuals. Full HBV genomes were recovered from these samples, two of which were from the Neolithic period, dating to about 7.000 and 5.000 years ago, and one from the medieval period. For the scientists it appears that the ancient virus genomes represent distinct lineages that have no close relatives today and possibly went extinct.
The two Neolithic genomes, although recovered from individuals that lived 2.000 years apart, were relatively similar to each other in comparison with modern strains, and were in fact more closely aligned with modern non-human primate strains of HBV than with modern human strains. In contrast, the medieval HBV genome is more similar to modern strains, but still represents a separate lineage. This is the case even when it is compared to two previously published HBV genomes recovered from mummies dating to the 16th century. The HBV strains found in these mummies are closely related to modern strains, suggesting a surprising lack of change in the virus over the last 500 years. These findings point to a complicated history for the virus, which may have involved multiple cross-species transmission events.
“Taken together, our results demonstrate that HBV already existed in Europeans 7.000 years ago and that its genomic structure closely resembled that of modern hepatitis B viruses, despite the differences observed,” explains first author Ben Krause-Kyora, junior professor at the IKMB. “More ancient precursors, intermediates and modern strains of both human and non-human primate HBV strains need to be sequenced to disentangle the complex evolution of this virus”, he adds.
Genetic evidences of ancient diseases: studying the evolution of blood-borne viruses
“It is a general statement that health issues and the incidence of diseases were reciprocally linked with substantial transformations of human lifeways in prehistory, for instance due to changes in diet or higher risks of epidemics due to increasing population density and interaction with livestock animals. Thanks to modern methods, we are able to disentangle this by studying ancient human as well as pathogen genetic makeups”, explains professor Almut Nebel from the IKMB. Together with Ben Krause-Kyora, Nebel runs the subproject F4 “Tracing Infectious Diseases in Prehistoric Populations” of the Collaborative Research Centre 1266 “Scales of Transformation in Prehistoric and Archaic Societies” at Kiel University. Central aim of the project is to evaluate the role of infectious diseases with regard to substantial changes in demography and human-environmental interaction between 15.000 BCE to 1 BCE. The individuals from the Neolithic site of Sorsum and medieval Petersberg are part of a large-scale screening procedure of well-dated skeletal remains to trace bacterial and viral pathogens.
Ben Krause-Kyora, junior professor at the IKMB is enthusiastic about the outstanding research network at Kiel University and the collaboration with the MPI in Jena. “Since its initial stage at Kiel University, aDNA research provoked scientists from different fields to further explore its potential with regard to human history. New disciplinary branches such as proteomics enhance the toolkit of studying ancient diseases and human genomes on a solid archaeological and medical background, well demonstrated by the new results on HBV. Among others, working together with scholars from Jena means the gateway to jointly interpret complex issues still relevant today and to shape sustainable structures in this comparatively young scientific field.”
Johannes Krause, senior author and director of the Department of Archaeogenetics at the MPI in Jena, emphasizes the most important implication of the study. “Our results reveal the great potential of ancient DNA from human skeletons to allow us to study the evolution of blood-borne viruses. Previously, there was doubt as to whether we would ever be able to study these diseases directly in the past,” he explains. “We now have a powerful tool to explore the deep evolutionary history of viral diseases.”
Original publication
Ben Krause-Kyora et al.; "Neolithic and Medieval virus genomes reveal complex evolution of Hepatitis B"; eLife; 2018