Seeing how cells work as never before
Yale is adding microscopy that provides an unprecedented view of specimens whose thickness had thwarted close inspection
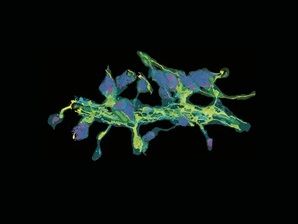
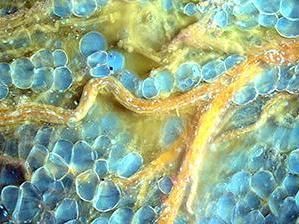
A neuronal dendrite captured by Yumei Wu, associate research scientist. Detail from FIB-SEM includes endoplasmic reticulum (yellow), mitochondria (green), and synaptic clefts (magenta).
Yumei Wu and Pietro De Camilli
Biological structures exist and function in three dimensions, but the limitations of imaging technology have long meant that scientists could examine only two dimensions in fine detail. The traditional practice of analyzing specimens one thin slice at a time, even if the structure being investigated is considerably thicker, can only get a researcher so far, says Derek K. Toomre, PhD, associate professor of cell biology, and director of the Yale “CINEMA” (Cellular Imaging using New Microscopy Approaches) laboratory. “Imagine if I had to recognize your face if I had just a slice through your skull,” he says. “It would be very hard.”
This summer, Yale is on track to install what for Toomre and his colleagues is a major advance: the s (FIB-SEM), which acquires tomography data from biological specimens. “There are relatively few of these microscopes around,” he notes. “There is not one in the region. It’s new and it’s something we’ve been wanting for a while, and I think it plays to an inherent strength of Yale, which has been in imaging.” For the first time, the z axis, which represents a specimen’s thickness, appears with resolutions that match those of two-dimensional x and y. FIB-SEM accomplishes this by imaging the surface of a thick cellular specimen (typically cells or tissue embedded in plastic or vitrified), and then using its ion beam to mill and etch away a very thin layer of the specimen while not damaging the remaining tissue. That exposes a new surface to be scanned. The process repeats, a few nanometers at a time. “And you can do that, drill down, as deep as you want,” says Toomre.
What emerges, layer by layer, is a computer-generated 3D image of the entire cell and all of its structures, which can be rotated to show views of the cell’s working parts from any angle. Pietro De Camilli, MD, chair and John Klingenstein Professor of Neuroscience and professor of cell biology, says FIB-SEM gives him unprecedented views of large volumes of neuronal cells and their specialized compartments, such as dendritic spines and axon terminals, including subtle details of the endoplasmic reticulum (ER), mitochondria, synaptic vesicles, and other organelles. “There is not a technique that allowed us to reconstruct with such a degree of precision the intracellular structure of a neuron, or any cell for that matter,” De Camilli says. “This technique is a fantastic tool for a cell biologist.”
Shirin Bahmanyar, PhD, assistant professor of molecular, cellular, and developmental biology, is one such scientist. Her work on cells’ nuclear envelope pockets was one example cited when De Camilli, Toomre, and Xinran Liu, MD, PhD, director of the electron microscopy facility in the Center for Cellular & Molecular Imaging, secured National Science Foundation funding to bring FIB-SEM to Yale. Bahmanyar is investigating how the DNA housed in the nuclear envelope of every cell in a living organism is accurately replicated, with the copy then safely encased when cells divide. It is an intricate process through which the ER—a complex network of membranes that generate the nuclear envelope—changes shape, opens and closes, and even appears to change its biochemical composition as division occurs, all for the purpose of preserving and propagating an organism’s most fundamental feature, its genetic code.
Bahmanyar wants to know what happens both when the process works well and when things go wrong. “If this process goes awry, and you don’t generate a single nuclear compartment, and you have chromosomes that are not enclosed in the single compartment, the genome is misregulated.” Resulting mutations, she says, can create malignant “supercells” that propagate better and faster than healthy cells and give cancers a biological advantage. She explains that with FIB-SEM, “we can see at an ultrastructure level how these compartments are wrapping around the nucleus, and how the chromosomes are stacked so that they stay together and generate this high-fidelity process that is essential to life. Essentially it would be impossible to look at this process given the large volume of a mitotic [dividing] cell, without this sort of microscopy.”
FIB-SEM can streamline the imaging process, letting investigators obtain fine structural detail of whole cells all within one imaging session. Liu, who will host and oversee the operation of the system, says a researcher can set various resolutions for different parts of the cell, and the system will be able to etch with the ion beam and then scan with electrons to form images automatically. “You can just set it up, and the process can be continued from a few hours to several days until a 3D dataset is obtained.” With FIB-SEM physically on campus, Yale investigators will be able to test far more of their hypotheses as well. The handful of similar instruments elsewhere, such as the Howard Hughes Medical Institute’s setup at its Janelia Research Campus in Virginia, are heavily subscribed. De Camilli says he has obtained striking images in Virginia, but at the same time, “these machines are terribly overbooked. We have been able to view a few specimens, but there is no way we can do something systematic without an instrument at Yale.”
Bahmanyar relishes the access she will have to Yale’s FIB-SEM while still a junior investigator. “Proximity, availability, all of those things just lower the barrier to new discoveries,” she says. “You can try things. You can get the people in your lab to go there and get trained themselves. To use the newest technologies is just so much faster and easily achievable if you have it at home.”
The technological strides represented at Yale by FIB-SEM and other modern imaging systems inspire Toomre to recall how prolific a giant of the past was with more primitive tools. The late George E. Palade, MD, Nobel Laureate and longtime School of Medicine faculty member, is widely considered the father of modern cell biology. One extraordinary aspect of Palade’s work, Toomre notes, is that he managed to define organelles and structures in cells despite seeing less than 1 percent of the volume of an individual cell in any given image. Just imagine today’s possibilities, Toomre says. “To see the cell in its entirety, to see tissues in three dimensions, in physiological and pathological conditions, the insights are much more profound. That is the revolution that we’ve been seeing with these volumetric approaches as will now be enabled at the medical school. They will be cutting edge with FIB-SEM.”