Illuminating the genome
RNA-guided endonuclease - in situ labelling, a new CRISPR/Cas9 based molecular visualisation method
The CRISPR/Cas9 system has been making ripples in the scientific community ever since its mechanisms were proposed in 2012. Commonly referred to as a genome editing tool, many scientists have found different applications for the scissor-like properties of the Cas9-protein. Researchers from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK Gatersleben) have now found a way to utilise the RNA/protein complex in a slightly different way - as a cytogenetic torch. Other than in conventional in situ hybridisation, the new RNA-guided endonuclease - in situ labelling-tool (RGEN-ISL) no longer requires denaturation of the DNA. The new method therefore leaves the chromatin intact, enabling investigation of the structure of the sample. Moreover, RGEN-ISL can be combined with protein-detection methods and allows real-time visualisation of the labelling-process. Whilst initially developed for plant genomes, RGEN-ISL can be used in all organisms and shows to be a promising new tool in the field of chromosome biology.
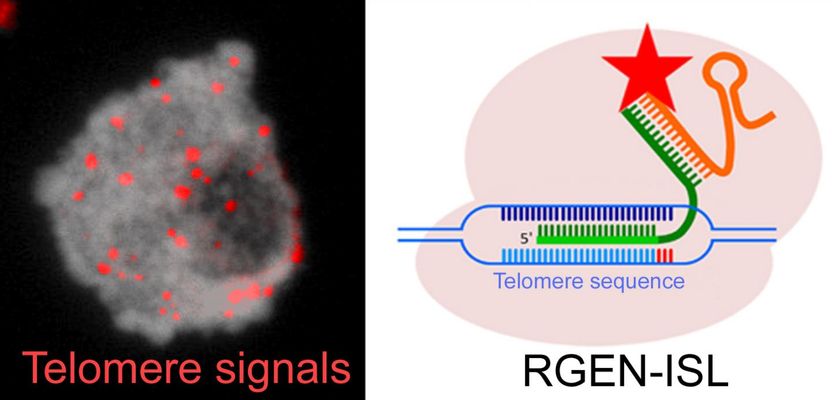
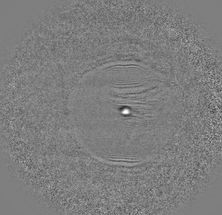
The nucleus of cowpea exhibiting telomere-specific signals (in red). The CRISPR/Cas9-based REGN-ISL method was used for the detection of telomeres.
Takayoshi Ishii, Andreas Houben
The discovery of the type II clustered regularly interspaced short palindromic repeats (CRISPR)-associated caspase 9 (Cas9) system has been a milestone in the field of targeted genome editing. Initially derived from the bacterium Streptococcus pyogenes, the RNA/protein-complex now is an established tool for targeted genome-editing in eukaryotes.
Whilst its scissor-like properties have yielded a broad range of applications, scientists from the Leibniz Institute of Plant Genetics and Crop Plant Research (IPK Gatersleben) are now using CRISPR/Cas9 in a new cytogenetic method for shining a light into eukaryote genomes - RNA-guided endonuclease - in situ labelling (RGEN-ISL).
For the last 30 years, Fluorescence in situ hybridization (FISH) has been the established and commonly used method for visualising in-situ DNA-sequences at chromosomal level. However, this method requires denaturation of the investigated DNA, thus often damaging the structure of the sample. By basing the RGEN-ISL method on CRISPR-Cas9, IPK researchers managed to bypass the denaturation-step of FISH whilst simultaneously integrating the desired fluorescent labelling properties of the conventional FISH-method. As the new cytogenetic tool preserves the structure of the sample, it opens up the option of investigating the spatio-temporal structure of the genome.
Further experimentation showed that RGEN-ISL outperforms conventional method combinations, such as FISH and immunohistochemistry, requiring less preparation and being comparatively quicker and cheaper. Additionally, the new method functions over a broad temperature-range of 4°C to 37°C and can also be combined with additional protein-detection and imaging methods. A further merit is that RGEN-ISL allows for real-time visualisation of the CRISPR/Cas9-mediated DNA-labelling, therefore revealing the kinetics of the reaction.
So far, the researchers have tested RGEN-ISL in plant samples, but also in human chromosomes, illustrating that their new method can likely be applied within all organisms. Currently, the use of method is limited to repetitive DNA-sequences, as often found in plant genomes. However, Dr Takayoshi Ishii, now working at Tottori University (Japan), who conceived the initial idea behind RGEN-ISL, assumes that this method could be adapted to visualise even single-copy sequences in the future.
Original publication
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.