Biologists develop new method of cloning
High efficiency at low cost
DNA, which contains the genetic information of an organism, consists of long “chains” of nucleotides. In order to study the functions based on the sequence of these building blocks, DNA molecules must be inserted in carrier molecules (plasmid-vectors) to be multiplied. For this cloning process, a research team from the University of Bayreuth has developed a highly efficient, fast and inexpensive method, versatile enough to be deployed in all areas of biology, biochemistry and biotechnology. A key feature of the method is that it makes any painstaking screening of bacterial colonies unnecessary.
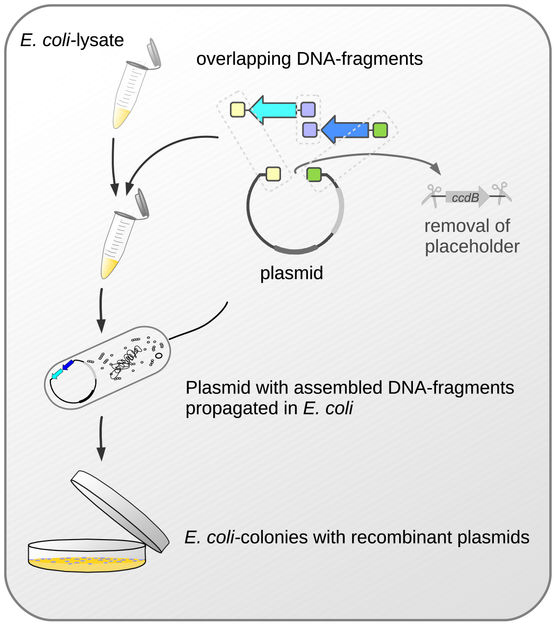
Important steps in the new cloning method.
David Oliver Richter
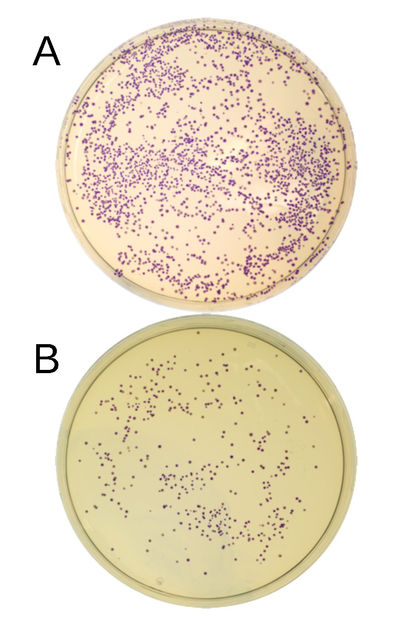
E. coli bacteria in the Petri dish, containing the recombinant plasmid (i.e. loaded with DNA fragments) and a producing blue protein. The blue protein coding DNA was assembled from two DNA fragments (A), or three DNA fragments (B). Each dot represents a bacterial colony. The number of colonies provides an insight into the efficiency of cloning.
David Oliver Richter
One thing that all methods subsumed under the term molecular cloning have in common is that the DNA fragments-of-interest are first incorporated into larger carrier molecules, the plasmid-vectors. Bacteria are then made to take up these vectors bearing the DNA-fragments. By the bacteria reproducing and forming bacterial colonies, the DNA fragments are multiplied thousands of times. Up to now, however, these methods have had one considerable drawback: since the insertion of DNA fragments into the carrier molecule does not always proceed as smoothly and perfectly as required, only some, but by no means all, colonies come to possess the vectors with the DNA fragments to be duplicated. In order to identify these “success stories”, time-consuming and expensive screening, up to now, has been unavoidable.
The Bayreuth researchers led by Prof. Dr. Stefan Schuster have now succeeded in making this screening redundant. The vector they have been utilizing is a plasmid that contains a toxic gene. DNA fragments are then incorporated into the plasmid in such a way as to replace this very gene. If this does not succeed, the plasmid keeps its toxic potential. And if, in turn, this plasmid is taken up by an E. coli-bacterium, its toxic effect sets in: It interferes with the bacterium’s cell division and therefore its ability to build colonies. In this way, it can be guaranteed right from the start, that only those E. coli-bacteria that do, in fact, contain the DNA fragments will form colonies. They will not subsequently have to be painstakingly selected. “The reason why our new cloning system is so efficient is that the selection of the bacteria equipped with the cloned DNA occurs reliably and all by itself. The multiplied plasmids can then be isolated from these bacteria and utilized – to analyse the function of the cloned DNA, or as the case may be, deployed in the biotechnological production of proteins”, says Bayreuth biologist Dr. David Richter, lead author of the study.
Moreover, the method presented in Scientific Reports simplifies the cloning procedure in one other aspect: The scientists also optimised an extract (SLiCE) derived from the cells of E. coli-bacteria to make it particularly suitable as a “glue” to string together several DNA fragments. Consequently, it is now possible to insert all sorts of combinations of DNA fragments into the plasmid – and that much more quickly than with previous methods.
The Bayreuth research team have named their new cloning system “ZeBRα”; the acronym deriving from the scientific terms for two decisive factors in their work. The plasmid used is a “Zero-Background Vector”. This means: Bacteria which do not contain the DNA fragments to be duplicated do not form inconvenient colonies in the background. Meanwhile “Redα-Exonuclease” is a component of the E. coli extract, with which various DNA fragments can be strung together and incorporated into the vector.
Building on the research results they have now published, the scientists intend to equip their cloning vectors with additional functions in the future to make it even more useful. In particular, they plan to further optimize the vector to be able to simplify the transformation of certain organisms or cell lines. Because such a transfer is also a rare event, it is advantageous if the vector also transports DNA sequences that lead to the formation of fluorescent proteins to monitor the process of transformation. These proteins will then indicate the successful uptake of the plasmid bearing DNA fragments into the organisms or the cells respectively.