All in a flash! Eliminate proteins from cells with a flash of light
LMU scientists have developed a tool to eliminate essential proteins from cells with a flash of light. The new method makes it possible to study the function of essential proteins.
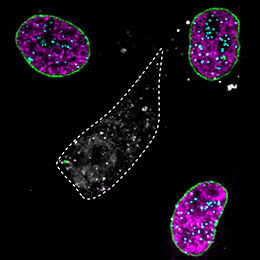
In the middle cell (grey), three different proteins were specifically removed: a protein of the nuclear envelope (Lamin A, green), a chromatin protein (CENPA, turquoise) and the replication protein (PCNA, magenta).
Dr. Wen Deng, LMU
Proteins not only provide much of the structural architecture of cells, they also perform most of the executive functions by acting as highly specific chemical catalysts. Hence, they are intimately involved in all fundamental biological processes, including metabolism, growth and cell division. Conversely, alterations in their shapes and activity result in the development of disorders. In order to understand the processes controlled by proteins, it is necessary to understand how each of them works. Biologists normally deduce the role of a protein by analyzing what happens when it is damaged or deleted altogether. Experimentally, this is usually accomplished by mutating or deleting the gene that encodes it. However, in the case of proteins that are essential for the survival of the organism, or of cell types that are required for a particular process, this approach is not very informative, as such mutants tend to die before it can provide insights into the protein’s actual function. Researchers led by Professor Heinrich Leonhardt at LMU‘s Biocenter have now developed a tool that gets around this problem. Their method makes use of either light or specific chemicals to trigger the selective degradation of the protein of interest. The procedure, and a review of the results so far obtained with it, are reported in the journal Nature Communications.
“In order to develop this technology, we reprogrammed the cell’s waste-disposal system,” says Leonhardt. Proteins that are defective or are no longer required are normally designated for disposal by enzymatically attaching a molecule called ubiquitin to them. The ubiquitin marker is then recognized by a molecular complex called a proteasome, which essentially acts as a shredder of the tagged proteins. The system developed by Leonhardt and his colleagues makes use of small antibodies known as nanobodies to target the protein of interest. The nanobody then brings a specialized ligase for the attachment of the ubiquitin marker – and the target protein is consigned to the proteasome for destruction.
In order to be able to control whether and when a target molecule is labelled and degraded, the scientists have incorporated an additional switch. The target molecule is only marked when the scientists activate this switch with light or small chemical molecules. “In this way, we can regulate the levels of proteins continuously. It’s just like using a dimmer, we can set the desired concentration of any given protein and observe the effects on cellular processes,” Leonhardt explains.
The LMU researchers first used the system to investigate a protein that plays a critical role in DNA replication. As it is essential for viability it cannot be genetically disabled. “We used the new method to temporarily deplete this protein in cells, and were able to elucidate its involvement in DNA repair,” says Wen Deng, lead author of the new study. The results showed that the protein acts as a central scaffold, to load other DNA repair proteins and efficiently fix the DNA damage.
One other advantage of the new tool is that it can be employed in whole organisms. In cooperation with the research group of Professor Barbara Conradt the researchers used the system to study cell death in in the nematode worm Caenorhabditis elegans, which is an established model organism in biology. At certain stages in the development of this worm, one of the two daughter cells produced by specific cell divisions is specifically eliminated. “By depleting the executioner protein with the aid of the new system, we were able to prevent the loss of the corresponding daughter cell and gain insights into the process of programmed cell death,” says Leonhardt.
Since the ubiquitin-proteasome system is found not only in all higher organisms, but also in Archaea and even in some bacteria, the team assumes that their approach to the targeted regulation of protein degradation will be broadly applicable. It could therefore make an important contribution to efforts to understand the functions of vital proteins.