An efficient tool to link X-ray experiments and ab initio theory
Results can even be calculated during the experiment
The electronic structure of complex molecules and their chemical reactivity can be assessed by the method of resonant inelastic x-ray scattering (RIXS) at BESSY II. However, the evaluation of RIXS data has so far required very long computing times. A team at BESSY II has now developed a new simulation method that greatly accelerates this evaluation. The results can even be calculated during the experiment. Guest users could use the procedure like a black box.
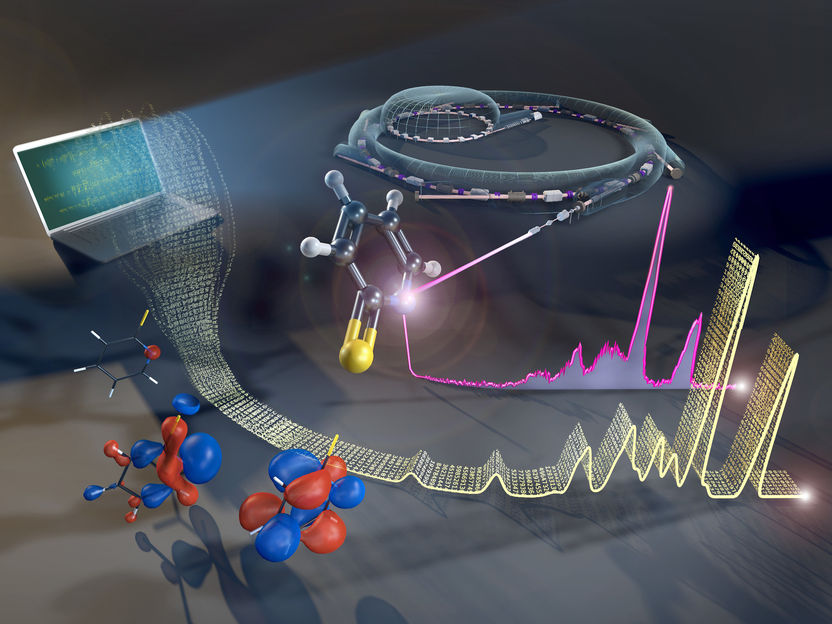
The electronic structure of complex molecules can be assessed by the method of resonant inelastic X-ray scattering (RIXS) at BESSY II.
© Martin Künsting /HZB
Molecules consisting of many atoms are complex structures. The outer electrons are distributed among the different orbitals, and their shape and occupation determine the chemical behaviour and reactivity of the molecule. The configuration of these orbitals can be analysed experimentally. Synchrotron sources such as BESSY II provide a method for this purpose: Resonant inelastic X-ray scattering (RIXS). However, to obtain information about the orbitals from experimental data, quantum chemical simulations are necessary. Typical computing times for larger molecules take weeks, even on high-performance computers.
Speeding up the evaluation
"Up to now, these calculations have mostly been carried out subsequent to the measurements", explains theoretical chemist Dr. Vinicius Vaz da Cruz, postdoc in Prof. Dr Alexander Föhlisch's team. Together with the RIXS expert Dr. Sebastian Eckert, also a postdoc in Föhlisch's team, they have developed a sophisticated new procedure that speeds up the evaluation many times over.
"With our method, it takes a few minutes and we don't need a super-computer for this, it works on desktop machines," says Eckert. The HZB scientists have tested the method on the molecule 2-thiopyridone, a model system for proton transfer, which are essential processes in living cells and organisms. Despite the short computing time, the results are precise enough to be very useful.
"This is a huge step forward," emphasises Föhlisch. "We can run through many options in advance and get to know the molecule, so to speak. In addition, this method also makes it possible to simulate far more complex molecules and to interpret the experimentally obtained data in a meaningful way". Experimental physicist Eckert adds: "We can now also run the simulations during the measurement and see immediately where it might be particularly exciting to take a closer look”.
The procedure is an extension of the well established and highly efficient time-dependent density functional theory, which is much faster than the traditional concepts to simulate the RIXS process. "The simplicity of the method allows for a large degree of automatization," says Vaz da Cruz: "It can be used like a black box."
Original publication
Most read news
Original publication
Vinıcius Vaz da Cruz, Sebastian Eckert and Alexander Föhlisch; "TD-DFT simulations of K-edge resonant inelastic X-ray scattering within the restricted subspace approximation"; Physical Chemistry Chemical Physics, issue 3, 2021
Topics
Organizations
Other news from the department science

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.