New capabilities in single-molecule sensing
DNA origami puts a smart lid on solid-state nanopore sensors
The latest advance in solid-state nanopore sensors – devices that are made with standard tools of the semiconductor industry yet can offer single-molecule sensitivity for label-free protein screening – expands their bag of tricks through bionanotechnology. Researchers at the Technische Universitaet Muenchen have enhanced the capabilities of solid-state nanopores by fitting them with cover plates made of DNA. These nanoscale cover plates, with central apertures tailored to various "gatekeeper" functions, are formed by so-called DNA origami – the art of programming strands of DNA to fold into custom-designed structures with specified chemical properties. The results are published in Angewandte Chemie International Edition.

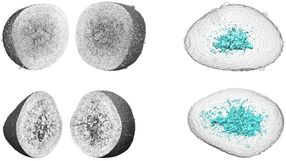
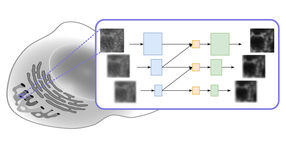
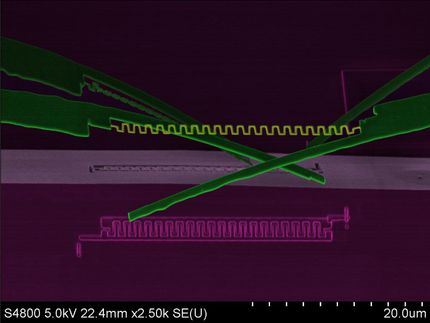
This illustration shows how a DNA origami nanoplate with a central aperture can serve as a smart lid or "gatekeeper" for a solid-state nanopore sensor. Researchers at the Technische Universitaet Muenchen have demonstrated that this arrangement can be used to filter biomolecules by size or to "fish" for specific target molecules by placing single-strand DNA receptors inside the aperture as "bait." With further research, they suggest, it might be possible to use such single-molecule sensors as the basis of a novel DNA sequencing system.
TU München, Hendrik Dietz and Ulrich Rant
Over the past few years, Prof. Hendrik Dietz's research group at TUM has been refining control over DNA origami techniques and demonstrating how structures made in this way can enable scientific investigations in diverse fields. Meanwhile, Dr. Ulrich Rant's research group has been doing the same for solid-state nanopore sensors, where the basic working principle is to urge biomolecules of interest, one at a time, through a nanometer-scale hole in a thin slab of semiconductor material. When biomolecules pass through or linger in such a sensor, minute changes in electrical current flowing through the nanopore translate into information about their identity and physical properties. Now Dietz and Rant, who are both Fellows of the TUM Institute for Advanced Study, have begun to explore what these two technologies can accomplish together.
The new device concept – purely hypothetical before this series of experiments – begins with the placement of a DNA origami "nanoplate" over the narrow end of a conically tapered solid-state nanopore. "Tuning" the size of the central aperture in the DNA nanoplate should allow filtering of molecules by size. A further refinement, placing single-stranded DNA receptors in the aperture as "bait," should allow sequence-specific detection of "prey" molecules. Conceivable applications include biomolecular interaction screens and detection of DNA sequences. In principle, such a device could even serve as the basis of a novel DNA sequencing system.
Step by step, the researchers investigated each of these ideas. They were able to confirm the self-assembly of custom-designed DNA origami nanoplates, and then their placement – after being electrically guided into position – over solid-state nanopores. They were able to demonstrate both size-based filtering of biomolecules and bait/prey detection of specific target molecules. "We're especially excited about the selective potential of the bait/prey approach to single-molecule sensing," Dietz says, "because many different chemical components beyond DNA could be attached to the appropriate site on a DNA nanoplate."
High-resolution sensing applications such as DNA sequencing will face some additional hurdles, however, as Rant explains: "By design, the nanopores and their DNA origami gatekeepers allow small ions to pass through. For some conceivable applications, that becomes an unwanted leakage current that would have to be reduced, along with the magnitude of current fluctuations."
Original publication
Other news from the department science

Get the analytics and lab tech industry in your inbox
From now on, don't miss a thing: Our newsletter for analytics and lab technology brings you up to date every Tuesday. The latest industry news, product highlights and innovations - compact and easy to understand in your inbox. Researched by us so you don't have to.


![[Fe]-hydrogenase catalysis visualized using para-hydrogen-enhanced nuclear magnetic resonance spectroscopy](https://img.chemie.de/Portal/News/675fd46b9b54f_sBuG8s4sS.png?tr=w-712,h-534,cm-extract,x-0,y-16:n-xl)