New tool for studying membrane protein structure
Membrane proteins are responsible for transporting chemicals and messages between a cell and its environment. But determining their structure has proved challenging for scientists. A study by UC Santa Barbara's Han Research Group demonstrates a new tool to resolve the structure of membrane-embedded and membrane-associating proteins using the water dynamics gradient they found across and above the lipid bilayer as a unique ruler.
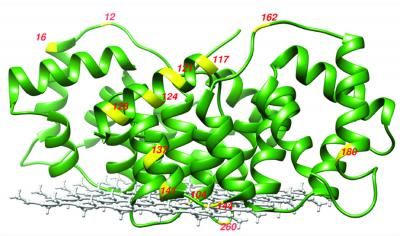
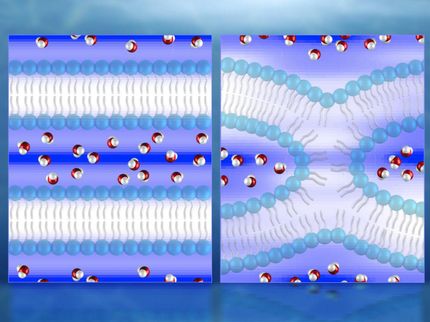
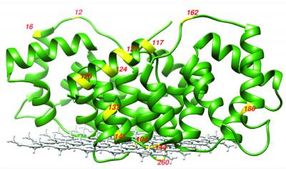
This is an X-ray crystal structure of membrane-bound annexing B12 in the presence of Ca2+. The residues studied are labeled in yellow.
UCSB
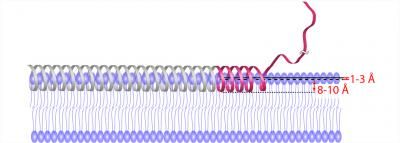
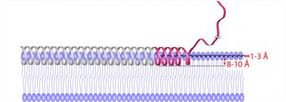
This is a structural representation of topology and immersion depth of membrane-bound alpha synuclein. The red ribbon is the topology obtained from study measurements; the gray ribbon represents observations from the literature.
UCSB
More than 25 percent of all human proteins are membrane proteins, which perform other essential functions, such as sensing and signaling. They also constitute approximately 50 percent of current drug targets, but the structure of only a small percentage has been resolved.
The UCSB study came about when researchers discovered that water on the membrane surface has a very distinct movement pattern: It is slowed down because the water is attracted to the membrane surface across several water layers. The scientists then wondered whether they could use this as an intrinsic ruler to determine how the associating protein is anchored into the membrane.
"It's very difficult to determine at what depth and in what conformation the protein is associating with the membrane, especially if you're talking about the interface or even the surface," said UCSB chemistry professor Song-I Han, the corresponding author of the study. "We found a contrast mechanism –– water dynamics –– which is distinctly different even above the membrane surface where there is no lipid density. The membrane surface distinctly changes the property of the water layers above it."
Postdoctoral scholar Chi-Yuan Cheng, the lead author of the study, and his colleagues used their unique spectroscopic tool to measure the water diffusivity at various positions of two membrane-associated proteins. These were carefully prepared by the Ralf Langen group at the University of Southern California, an expert team in the structure studies of membrane-associating proteins. The UCSB researchers then used the water dynamics gradient along the bilayer as an intrinsic ruler to determine structural information, such as topology, immersion depth and the location of the proteins, including the protein segments residing well away from the membrane surface –– information that was previously unresolved.
"Membrane proteins can sit deep inside the membrane but also associate at the periphery of the membrane," explained Cheng. "This can play a very important role in function, especially for peripheral and interfacial proteins."
"We are not suggesting our study can resolve the structure entirely, but it offers an important and missing puzzle piece to this problem," said Han. "While we may not be able to determine the entire structure, knowing the location and structure of a protein segment at the surface of membranes is very important."
The team used Overhauser dynamic nuclear polarization enhanced nuclear magnetic resonance (NMR), a technique they developed over the last few years. Using a small and stable radical with an even higher magnetic property than the hydrogen atom of water, researchers exploited this magnetic dipole by inserting it as a spin probe in the protein or membrane position of interest. They then used microwave irradiation to excite the dipole, which subsequently excited nearby water molecules, only when they moved at the same frequency as the dipole. In effect, the water near the spin probe was polarized, causing large NMR signal enhancements that could be measured to extract the local water dynamics.
"People have used this NMR relaxometry method before," said Han, "but what is novel is the fact the we use an electron spin as our excitation source, which has a much higher frequency than previously exploited, and that we actively drive the excitation of these spin probes with microwaves. The spin probes process at 10 gigahertz –– instead of hundreds of megahertz –– which allows us to look at the faster water dynamics relevant here as they are altered on biomolecular surfaces."
This proof of principal study is just the first step. Next the team will scrutinize neurodegenerative proteins, specifically tau, which plays a key role in Alzheimer's disease. The working hypothesis is that at some aggregation stages, tau may exert toxicity as it breaks through the membrane barrier, in part determined by its slowed surface water dynamics. The new tool developed by UCSB's Han Research Group can be used to test this hypothesis.
"You can imagine that the proof or disproof of the concept is hampered by the fact that it is very difficult to look at protein-membrane association," Han said. "Now we have a way of measuring these molecular assemblies. So if it's true that certain proteins species or their oligomers poke through, basically breaking down this barrier and making the membrane leakier, we surely should be able to see those features by looking at the surface water dynamics."