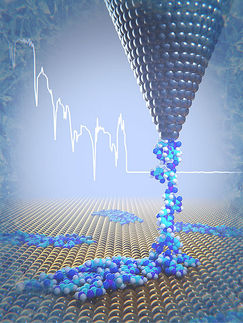
Measuring forces in the DNA molecule
First direct measurements of base-pair bonding strength
DNA, our genetic material, normally has the structure of a twisted rope ladder. Experts call this structure a double helix. Among other things, it is stabilized by stacking forces between base pairs. Scientists at the Technical University of Munich (TUM) have succeeded at measuring these forces for the very first time on the level of single base pairs. This new knowledge could help to construct precise molecular machines out of DNA.
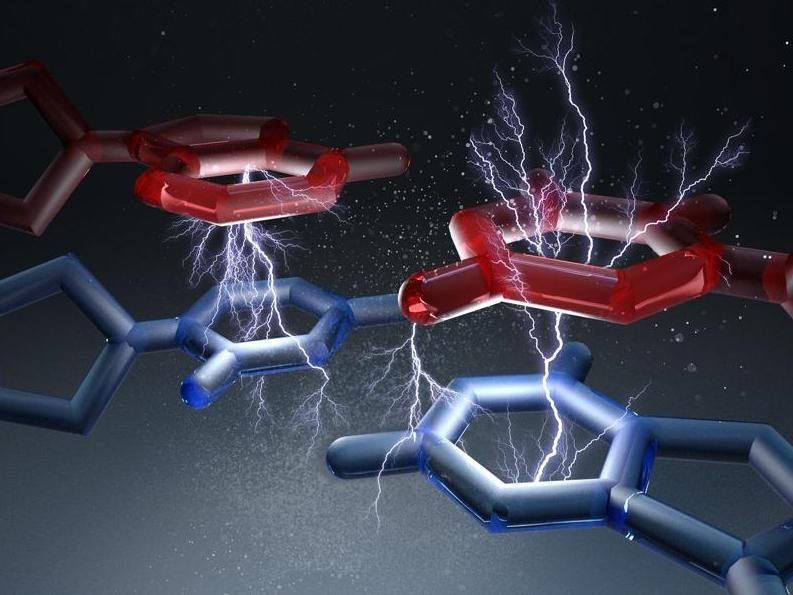
Illustration of base-pair bonding.
Christoph Hohmann & Hendrik Dietz/ Nano Initiative Munich/ TUM
Over 60 years ago, the researchers Crick and Watson identified the structure of deoxyribonucleic acid, which is more commonly known as DNA. They compared the double helix to a rope ladder that had been twisted into a spiral. The rungs of this ladder consisted of guanine/cytosine and thymine/adenine base pairs. But what keeps the DNA strands in that spiral structure?
Special measuring system for molecular interactions
Prof. Hendrik Dietz from the Chair of Experimental Biophysics uses DNA as construction material to create molecular structures. Hence, he is greatly interested in gaining a better understanding of this material. "There are two types of interactions which stabilize double helices", he explains. For one, DNA contains hydrogen bonds.
For another, there are what experts call base pair stacking forces, which act between the stacked base pairs along the spiral axis. The forces of the hydrogen bonds, on the other hand, act perpendicular to the axis. "So far, it is not quite clear to which extent these two forces each contribute to the overall stability of the DNA double helix”, explains Dietz.
Directly measuring the weak stacking forces between base pairs was a big technical challenge for the researchers, who worked on the problem for six years. In collaboration with the TUM Chair of Molecular Biophysics (Prof. Matthias Rief) and the TUM Chair of Theoretical Biophysics - Biomolecular Dynamics (Prof. Martin Zacharias), they succeeded in developing a special experimental setup that now makes it possible to measure extremely weak contact interactions between individual molecules.
A trillionth of a bar of chocolate
To put it simply, the measurement system is designed hierarchically and involves microscopic beams, at the tips of which one or more double helix structures running in parallel are located. These have been modified such that each end carries one base pair. Two of these microscopic beams are connected with a flexible polymer. On the other side, the beams are coupled to microscopic spheres which can be pulled apart using optical laser tweezers. In solution, the base pairs on the end of one of the beam can now interact with the base pairs on the end of the other beam. This also makes it possible to measure how long a stacking bond between them lasts before they fall apart again, as well as the force acting between the base pairs.
The forces measured by the researchers were in the range of piconewtons. "A newton is the weight of a bar of chocolate", explains Dietz. "What we have here is a thousandth of a billionth of that, which is practically nothing." Forces in the range of two piconewtons are sufficient to separate the bond created by stacking forces. Furthermore, the scientists also observed that the bonds spontaneously broke up and formed again within just a few milliseconds. The strength and the lifetime of the interactions depends to a great extent on which base pairs are stacked on each other.
Creating DNA machines
The results of the measurements may help to better understand mechanical aspects of fundamental biological processes such as DNA replication, i.e. the reproduction of genetic material. For example, the short life of the stacking interactions could mean that an enzyme tasked with separating the base pairs during this process just needs to wait for the stacking bonds break up on their own – instead of having to apply force to separate them.
However, Dietz also intends to apply the data directly to his current research: He uses DNA as programmable building material to construct machines on the order of nanometers. When doing so, he draws inspiration from the complex structures which can e.g. be found in cells and, among other things, serve as molecular "factories" to synthesize important compounds such as ATP, which stores energy. "We now know what would be possible if we could just build structures that were sufficiently sophisticated", says Dietz. "Naturally, when we have a better understanding of the properties of the molecular interactions, we are better able to work with these molecules."
At the moment, the lab is building a molecular rotational motor out of DNA, the components of which interlock and are held together via stacking forces. The goal is to be able to control a directed rotation via chemical or thermal stimuli.