Research team identifies structure of tumor-suppressing protein
An international group of researchers led by Carnegie Mellon University physicists Mathias Lösche and Frank Heinrich have established the structure of an important tumor suppressing protein, PTEN. Their findings provide new insights into how the protein regulates cell growth and how mutations in the gene that encodes the protein can lead to cancer. Phosphatase and tensin homolog (PTEN) is a known tumor suppressing protein that is encoded by the PTEN gene. When expressed normally, the protein acts as an enzyme at the cell membrane, instigating a complex biochemical reaction that regulates the cell cycle and prevents cells from growing or dividing in an unregulated fashion. Each cell in the body contains two copies of the PTEN gene, one inherited from each parent. When there is a mutation in one or both of the PTEN genes, it interferes with the protein's enzymatic activity and, as a result inhibits its tumor suppressing ability.
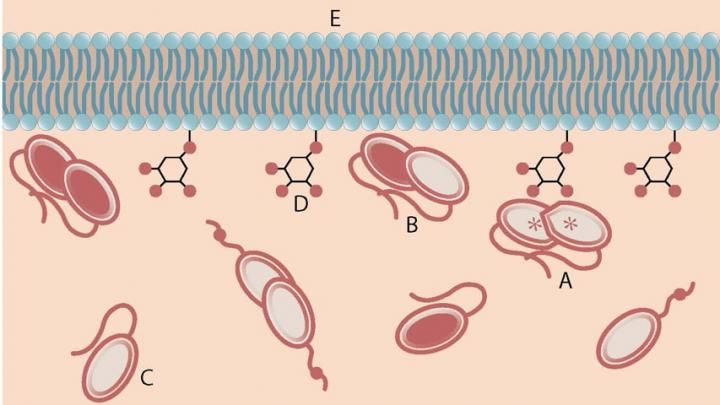
An activated PTEN dimer that contains two non-mutant proteins (A) can transform the functional lipid (D) on the cellular membrane (E) into a chemical form that tunes down cancer predilection. Dimers that contain a mutated protein (B), or PTEN monomers can not transform the functional lipid
Carnegie Mellon University
"Membrane-incorporated and membrane-associated proteins like PTEN make up one-third of all proteins in our body. Many important functions in health and disease depend on their proper functioning," said Lösche, who with other researchers within Carnegie Mellon's Center for Membrane Biology and Biophysics aim to understand the structure and function of cell membranes and membrane proteins. "Despite PTEN's importance in human physiology and disease, there is a critical lack of understanding of the complex mechanisms that govern its activity."
"PTEN dimerization may be the key to understanding an individual's susceptibility for PTEN-sensitive tumors," said Lösche, a professor of physics and biomedical engineering at Carnegie Mellon.
In order to reveal how dimerization improves PTEN's ability to thwart tumor development, researchers needed to establish the protein's dimeric structure. Normally, protein structure is identified using crystallography, but attempts to crystallize the PTEN dimer had failed. Lösche and colleagues used a different technique called small-angle X-ray scattering (SAXS) which gains information about a protein's structure by scattering X-rays through a solution containing the protein. They then used computer modeling to establish the dimer's structure.
They found that in the PTEN dimers, the C-terminal tails of the two proteins may bind the protein bodies in a cross-wise fashion, which makes them more stable. As a result, they can more efficiently interact with the cell membrane, regulate cell growth and suppress tumor formation.
Original publication

Get the analytics and lab tech industry in your inbox
By submitting this form you agree that LUMITOS AG will send you the newsletter(s) selected above by email. Your data will not be passed on to third parties. Your data will be stored and processed in accordance with our data protection regulations. LUMITOS may contact you by email for the purpose of advertising or market and opinion surveys. You can revoke your consent at any time without giving reasons to LUMITOS AG, Ernst-Augustin-Str. 2, 12489 Berlin, Germany or by e-mail at revoke@lumitos.com with effect for the future. In addition, each email contains a link to unsubscribe from the corresponding newsletter.